EZ Cap™ Firefly Luciferase mRNA
EZ Cap™ Firefly Luciferase mRNA will express luciferase protein once entering cells, which is initially extracted from firefly Photinus pyralis. This enzyme catalyzes ATP-dependent D-luciferin oxidation and lead to yield chemiluminescence at about 560 nm wavelength. Firefly Luciferase is a frequently used bioluminescent reporter for gene regulation and function study. It is applicable in assays for mRNA delivery, translation efficiency, cell viability and in vivo imaging etc.
Key features:
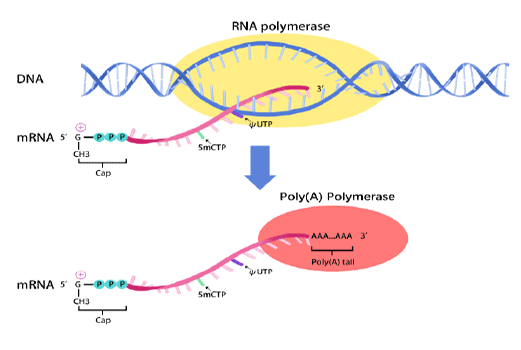
Cap1 Structure: The mRNA incorporates a Cap1 analog at the 5' end, ensuring high translation initiation efficiency, improved stability, and reduced innate immune recognition, leading to stronger and more sustained luciferase expression.
poly(A) tail: It contains an optimized poly(A) tail of approximately 100 nucleotides. This specific length is engineered to maximize transcript stability by resisting degradation and to synergize with the 5' cap for superior, sustained translation efficiency, ensuring robust protein yield in your applications.
| mRNA Length | 1921 nucleotides | ||
| Concentration | 1 mg/mL | ||
| Buffer | 1 mM Sodium Citrate, pH 6.4 | Storage | -40°C or below |
| General tips | Please dissolve it on ice and protect from RNase carefully. Avoid repeated freeze/thaw cycles as possible. Don’t vortex. Upon first use, centrifuge the tube softly and aliquot it into several single use portions. Use RNase-free reagents and materials with appropriate RNase-free technique. Don’t add to the media with serum unless mixing with a transfection reagent. | ||
| Shipping Condition | ship with dry ice. | ||
- 1. Shuling Ren, Xinyu Lin, et al. "Redox-Responsive Peptide Coacervates for Enhanced mRNA Delivery and Intracellular Release." ACS Nano. 2025 Dec 26. PMID: 41451635
- 2. Xu-Han Liu, Hui-Ping Song, et al. "Trehalose-loaded LNPs enhance mRNA stability and bridge in vitro in vivo efficacy gap." npj Vaccines 10, 201 (2025)
- 3. Caitlin McMillan, Amy Druschitz, et al. "Influence of ionisable lipid and sterol variations on lipid nanoparticle properties and performance." J Control Release. 2025 Jul 20:114056. PMID: 40695375
- 4. Dao Thi Hong Le, Chuan Yang, et al. "Replacing PEG-Lipid with Amphiphilic Polycarbonates in mRNA-Loaded Lipid Nanoparticles: Impact of Polycarbonate Structure on Physicochemical and Transfection Properties." Biomacromolecules. 2025 Jun 9;26(6):3563-3575. PMID: 40347133
- 5. Siyu Dong, Zhaoqi Pan, et al. "Lipid Nanoparticles - Mediated mRNA Delivery to the Eye Affected by Ionizable Cationic Lipid." Mol Pharm. 2025 Jun 2;22(6):3297-3307. PMID: 40331524
- 6. Xueying Tang, Jiashuo Zhang, et al. "Autonomic lysosomal escape via sialic acid modification enhances mRNA lipid nanoparticles to eradicate tumors and build humoral immune memory." J Control Release. 2025 Jun 10:382:113647. PMID: 40158813
- 7. Timothy H. Cheung, Alexander Fuchs, et al. "Acid-Responsive Polymer Additives Increase RNA Transfection from Lipid Nanoparticles." ADVANCED FUNCTIONAL MATERIALS Volume35, Issue2 January 9, 2025 2413220
- 8. Jingjiao Li, Jie Hu, et al. "High-throughput synthesis and optimization of ionizable lipids through A3 coupling for efficient mRNA delivery." J Nanobiotechnology. 2024 Nov 4;22(1):672. PMID: 39497197
- 9. Caitlin McMillan, Amy Druschitz, et al. "Tailoring lipid nanoparticle dimensions through manufacturing processes." RSC Pharm. 2024 Sep 23. PMID: 39323767
- 10. Kathryn A. Whitehead, et al. "Lipid nanoparticle structure and delivery route during pregnancy dictate mRNA potency, immunogenicity, and maternal and fetal outcomes." Proc Natl Acad Sci U S A. 2024 Mar 12;121(11):e2307810121. PMID: 38437545
- 11. Peng Zhang, Xiaoqing Hu, et al. "Schlafen-11 and -9 are innate immune sensors for intracellular single-stranded DNA." bioRxiv 28 February 2024
- 12. Yixuan Huang, Jiacai Wu, et al. "Quaternization drives spleen-to-lung tropism conversion for mRNA-loaded lipid-like nanoassemblies." Theranostics. 2024 Jan 1;14(2):830-842. PMID: 38169552
- 13. Yutong Hou, Sihao Lin, et al. "Alleviation of ischemia–reperfusion induced renal injury by chemically modified SOD2 mRNA delivered via lipid nanoparticles." Mol Ther Nucleic Acids. 2023 Oct 26:34:102067. PMID: 38028193
- 14. Mingzhu Gao, Maoping Tang, et al. "Modulating Plaque Inflammation via Targeted mRNA Nanoparticles for the Treatment of Atherosclerosis." ACS Nano. 2023 Sep 26;17(18):17721-17739. PMID: 37669404
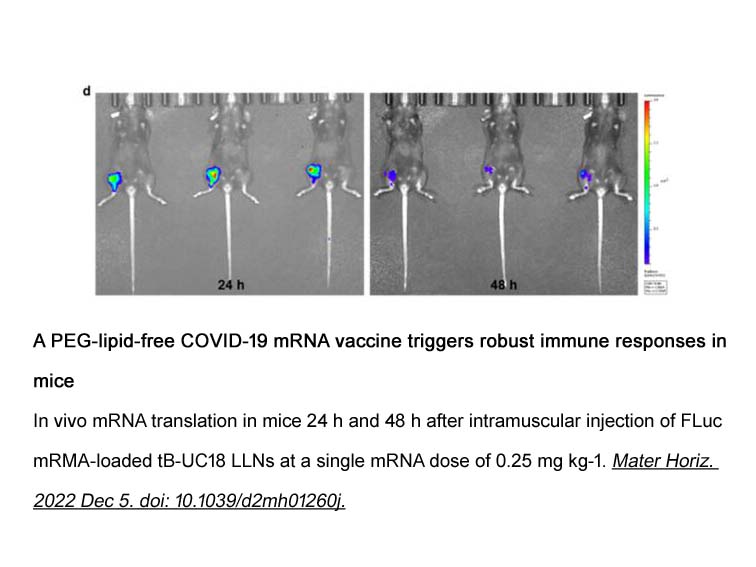
- 15. Min Li, Yixuan Huang, et al. "A PEG-lipid-free COVID-19 mRNA vaccine triggers robust immune responses in mice." Mater Horiz. 2023 Feb 6;10(2):466-472. PMID: 36468425
- 16. Shaoyan Gao, Xiaohe Li, et al. "PKM2 promotes pulmonary fibrosis by stabilizing TGF-β1 receptor I and enhancing TGF-β1 signaling." Sci Adv. 2022 Sep 23;8(38):eabo0987. PMID: 36129984
- 17. Y. Huang, M. Yang, et al. "Intracellular delivery of messenger RNA to macrophages with surfactant-derived lipid nanoparticles." Materials Today Advances Volume 16, December 2022, 100295
- 18. Jiaxing Di, Zhili Du, et al. "Biodistribution and Non-linear Gene Expression of mRNA LNPs Affected by Delivery Route and Particle Size." Pharm Res. 2022 Jan;39(1):105-114. PMID: 35080707
- 19. Min Li , Sanpeng Li, et al. "Secreted Expression of mRNA-Encoded Truncated ACE2 Variants for SARS-CoV-2 via Lipid-Like Nanoassemblies." Adv Mater. 2021 Aug;33(34):e2101707. PMID: 34278613
Quality Control & Datasheet
- View current batch:
Related Biological Data
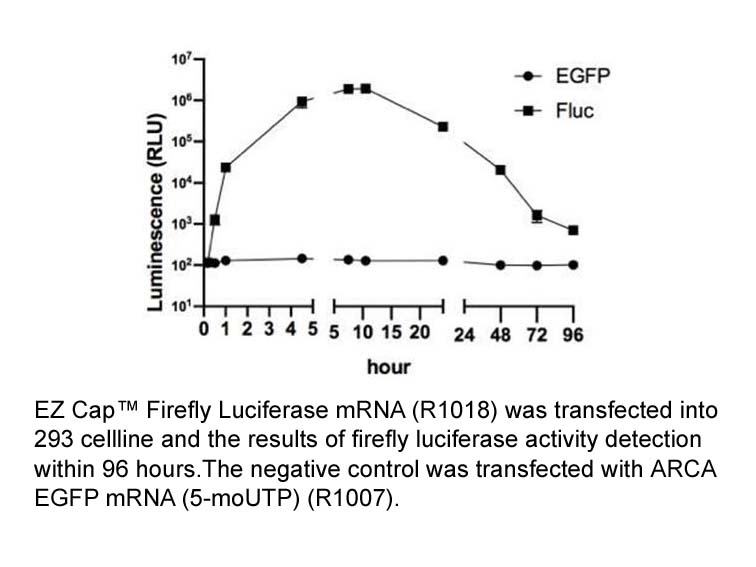
Related Biological Data
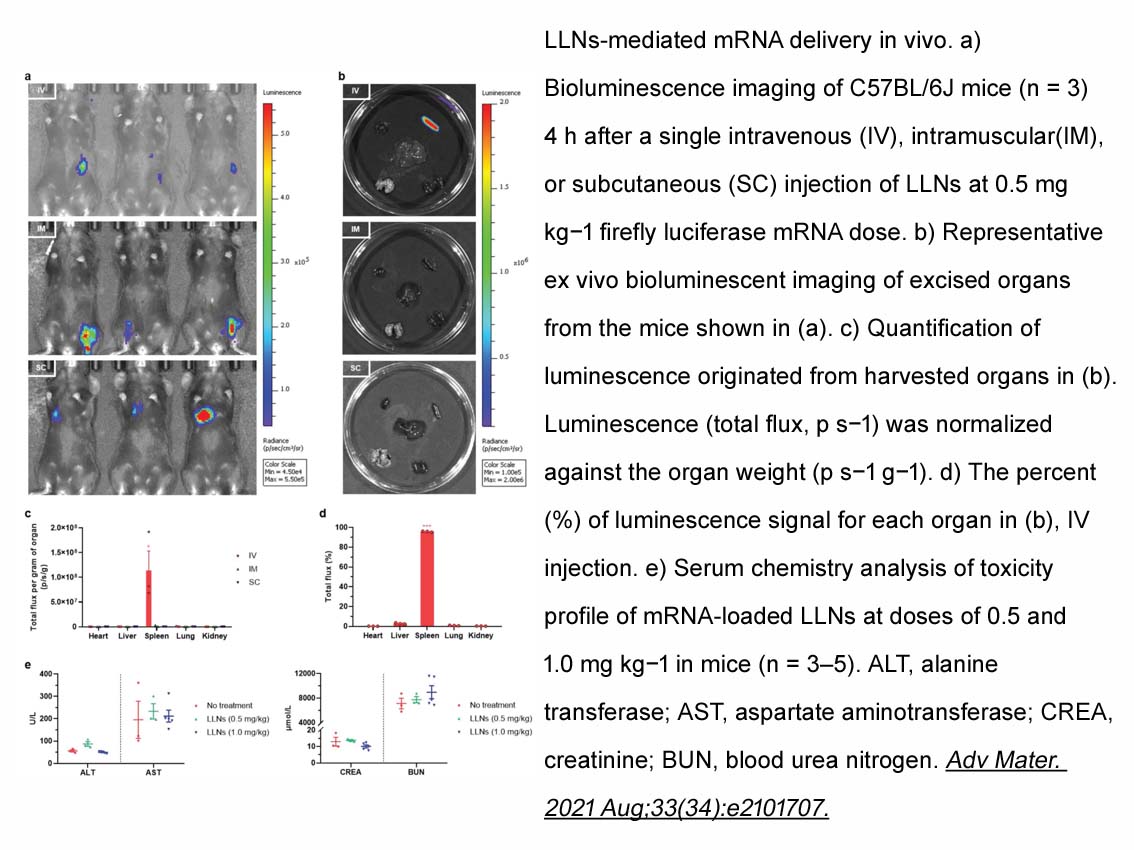
Related Biological Data
It can be based on your experimental goals:
- For Tracking Transfection and Translation Efficiency: APExBIO Reporter Gene mRNAs (e.g. EGFP, Firefly Luciferase mRNA) are commonly used to track transfection efficiency and protein expression duration; evaluate gene expression and cell viability; study mRNA localization and bio-distribution via in vivo imaging; optimize transfection conditions and validate LNP delivery system.
- For Gene Editing, Functional Studies and Gene Therapy Research: APExBIO offers various functional protein mRNAs, involving tumor suppressors (e.g. p53, PTEN), cytokines (e.g. IL-12, IL-10), gene-editing tools (e.g. spCas9, Cre Recombinase), gene replacement protein (e.g. EPO), and antigens (e.g. OVA, SARS-CoV-2 Spike).
- For Sustained Protein Expression: APExBIO Self-amplifying RNA (saRNA) and Circular RNA (circRNA) are recommended for applications requiring prolonged protein expression. saRNA enables lasting and strong protein expression at lower doses, while circRNA has enhanced structural stability and extended expression duration.
- Advanced Capping Technology: Utilizes Cap 1 structure (EZ Cap™ Cap) to achieve enhanced translation efficiency and minimizing activation of the host innate immune response. The capping efficiency can reach 90–99%.
- Diverse Modification Options: Provides a range of modified nucleotides, such as m1Ψ, 5-moUTP and Cy5-UTP, which reduce immunogenicity, improve mRNA stability, and maximize protein expression levels.
- Stringent Quality Control: Each batch undergoes rigorous quality assessment including capping efficiency, purity, integrity, and sterility to ensure batch-to-batch consistency.