THZ1
THZ1 is a covalent inhibitor of CDK7 with IC50 value of 3.2nM [1].
THZ1 covalently modifies CDK7 by targeting C312 residue outside of the kinase domain, providing an unanticipated means of achieving covalent selectivity. THZ1 potently inhibits proliferation of Jurkat and Loucy T-ALL cell lines with IC50 values of 50nM and 0.55nM, respectively. In the kinase binding assay, THZ1 shows a good binding affinity with IC50 value of 3.2nM [1].
As an inhibitor of CDK7, THZ1 inhibits the phosphorylation of the C-terminal domain of RNAP polymerase II, effecting the regulation of transcription. THZ1 also inhibits the activation of the CDK proteins. It is reported to disrupt the CDK7 signalling pathways both in Jurkat cells and Loucy cells. THZ1 shows a broad-based activity with IC50 values less than 200nM in a variety of cancer cell lines. Among these cell lines, T-ALL is exceptional sensitivity to THZ1 due to the transcription effect of RUNX1 caused by THZ1 [1].
References:
[1] Nicholas Kwiatkowski, Tinghu Zhang, Peter B. Rahl et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature, 2014.
- 1. Roberta Cacioppo, Alexander Gillis, et al. "CRL3ARMC5 ubiquitin ligase and Integrator phosphatase form parallel mechanisms to control early stages of RNA Pol II transcription." Article Volume 84, Issue 24, p4808-4823.e13, December 19, 2024.
- 2. Yi Sui, Teng Wang, et al. "Targeting Super-Enhancer-Driven Transcriptional Dependencies Suppresses Aberrant Hedgehog Pathway Activation and Overcomes Smoothened Inhibitor." Cancer Res. 2024 May 22. PMID: 38775809
- 3. Anh Phuong Nguyen, Kaoru Yamagata, et al. "Enhancer RNA commits osteogenesis via microRNA-3129 expression in human bone marrow-derived mesenchymal stem cells." Inflamm Regen. 2022 Sep 16;42(1):43. PMID: 36114571
- 4. Te Zhang, Xuming Song, et al. "Aberrant super-enhancer landscape reveals core transcriptional regulatory circuitry in lung adenocarcinoma." Oncogenesis. 2020 Oct 17;9(10):92. PMID: 33070167
- 5. Cho YS, Li S, et al. "CDK7 regulates organ size and tumor growth by safeguarding the Hippo pathway effector Yki/Yap/Taz in the nucleus." Genes Dev. 2020 Jan 1;34(1-2):53-71. PMID: 31857346
- 6. Maritere Urioistegui-Arcos, Rodrigo Aguayo-Ortiz, et al. "Disruption of TFIIH activities generates a stress gene expression response and reveals possible new targets against cancer." bioRxiv. 2019 December 03.
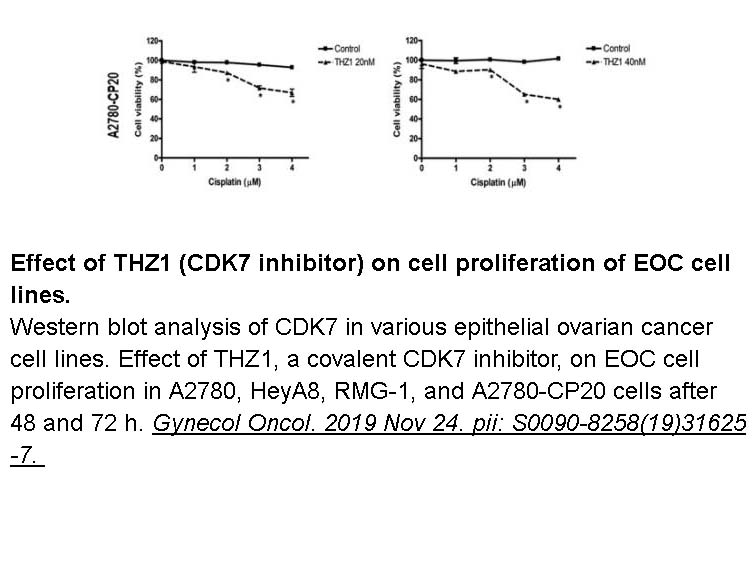
- 7. Kim J, Cho YJ, et al. "CDK7 is a reliable prognostic factor and novel therapeutic target in epithelial ovarian cancer." Gynecol Oncol. 2019 Nov 24. pii: S0090-8258(19)31625-7. PMID: 31776040
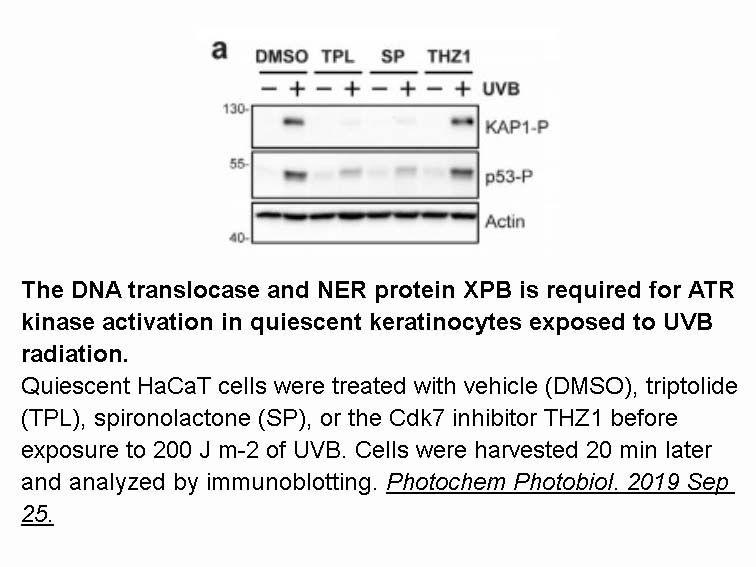
- 8. Shaj K, Hutcherson RJ, et al. "ATR Kinase Activity Limits Mutagenesis and Promotes the Clonogenic Survival of Quiescent Human Keratinocytes Exposed to UVB Radiation." Photochem Photobiol. 2019 Sep 25. PMID: 31554014
- 9. Sava GP, Fan H, et al. "ABC-transporter upregulation mediates resistance to the CDK7 inhibitors THZ1 and ICEC0942." Oncogene. 2019 Sep 17. PMID: 31530935
- 10. KAVYA SHAJ. "DIFFERING FUNCTIONS OF ATR KINASE IN HUMAN EPIDERMAL KERATINOCYTES EXPOSED TO ULTRAVIOLET B RADIATION." Wright State University. 2019.
- 11. Seoane M, Buhs S, et al. "Lineage-specific control of TFIIH by MITF determines transcriptional homeostasis and DNA repair." Oncogene. 2019 Jan 16. PMID: 30651597
- 12. Landsverk HB, Sandquist LE, et al. "Regulation of ATR activity via the RNA polymerase II associated factors CDC73 and PNUTS-PP1." Nucleic Acids Res. 2018 Dec 12. PMID: 30541148
- 13. Lin L, Huang M, et al. "Super-enhancer-associated MEIS1 promotes transcriptional dysregulation in Ewing sarcoma in co-operation with EWS-FLI1." Nucleic Acids Res. 2018 Nov 28. PMID: 30496486
- 14. Zanconato F, Battilana G, et al. "Transcriptional addiction in cancer cells is mediated by YAP/TAZ through BRD4." Nat Med. 2018 Oct;24(10):1599-1610. PMID: 30224758
- 15. Chen D, Zhao Z, et al. "Super enhancer inhibitors suppress MYC driven transcriptional amplification and tumor progression in osteosarcoma." Bone Res. 2018 Apr 4;6:11. PMID: 29644114
- 16. Helga B. Landsverka1, Lise E. Sandquista,et al. "Regulation of ATR activity by the RNA polymerase II phosphatase PNUTS-PP1." bioRxiv.2018.February 16. PMID: 29498802
- 17. Yuan J, Jiang YY, et al. "Super-Enhancers Promote Transcriptional Dysregulation in Nasopharyngeal Carcinoma." Cancer Res. 2017 Dec 1;77(23):6614-6626. PMID: 28951465
- 18. Kemp MG. "DNA damage-induced ATR kinase activation in non-replicating cells is regulated by the XPB subunit of transcription factor II-H (TFIIH). J Biol Chem." 2017 Jun 7. pii: jbc.M117.788406. PMID: 28592488
- 19. Liao R, Mizzen CA. "Site-specific regulation of histone H1 phosphorylation in pluripotent cell differentiation." Epigenetics Chromatin. 2017 May 22;10:29. PMID: 28539972
- 20. Francavilla C, Lupia M, et al. "Phosphoproteomics of Primary Cells Reveals Druggable Kinase Signatures in Ovarian Cancer." Cell Rep. 2017 Mar 28;18(13):3242-3256. PMID: 28355574
- 21. Posternak V, Ung MH, et al. "MYC Mediates mRNA Cap Methylation of Canonical Wnt/β-Catenin Signaling Transcripts By Recruiting CDK7 and RNA Methyltransferase." Mol Cancer Res. 2017 Feb;15(2):213-224. PMID: 27899423
- 22. Harrod A, Fulton J, et al. "Genomic modelling of the ESR1 Y537S mutation for evaluating function and new therapeutic approaches for metastatic breast cancer." Oncogene. 2017 Apr 20;36(16):2286-2296. PMID: 27748765
| Storage | Store at -20°C |
| M.Wt | 566.05 |
| Cas No. | 1604810-83-4 |
| Formula | C31H28ClN7O2 |
| Solubility | ≥28.3 mg/mL in DMSO; insoluble in H2O; insoluble in EtOH |
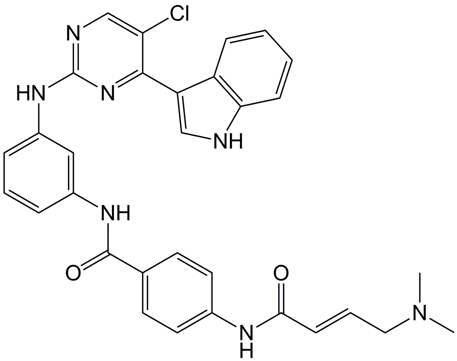
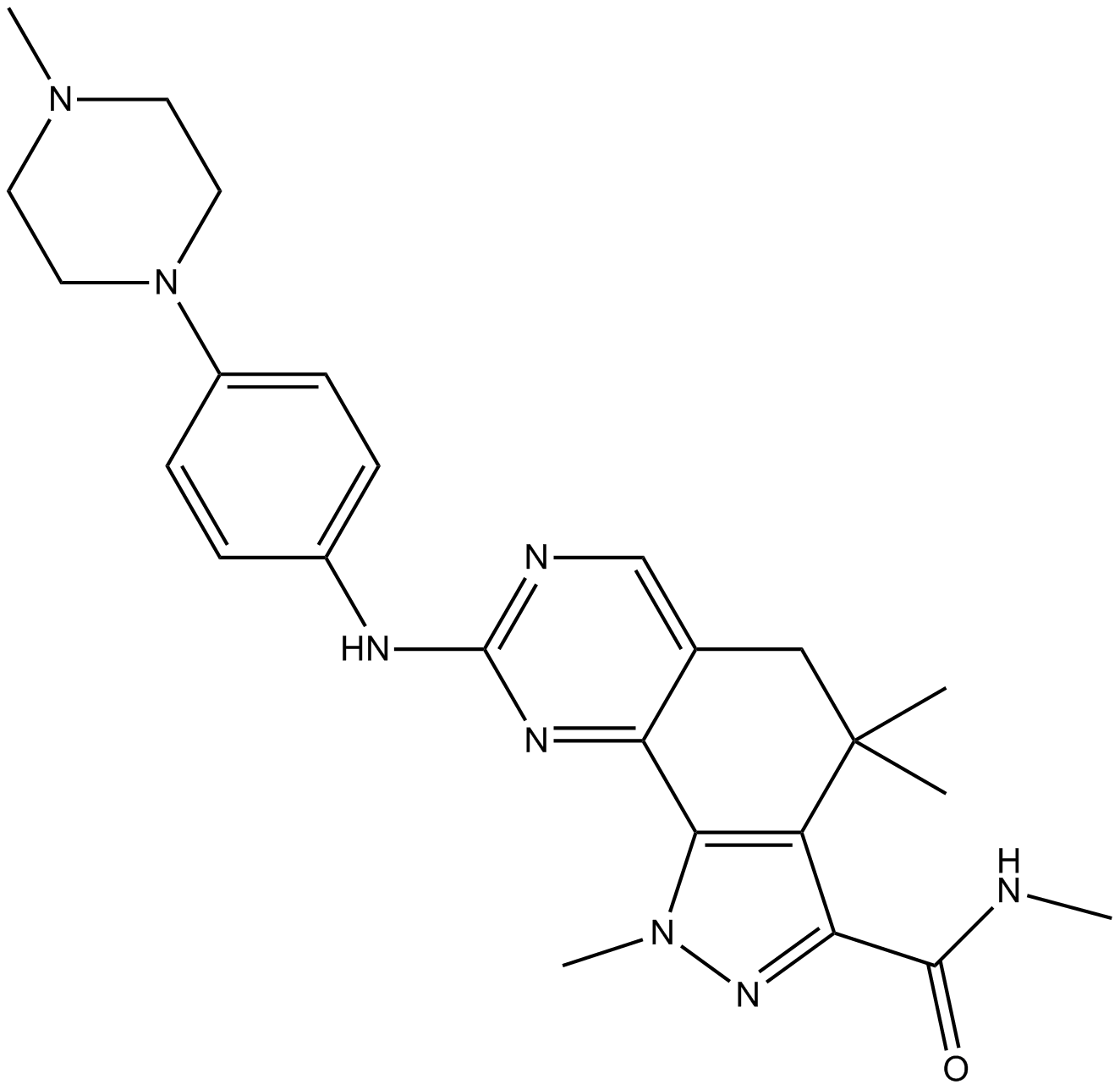
| Chemical Name | (E)-N-(3-((5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl)amino)phenyl)-4-(4-(dimethylamino)but-2-enamido)benzamide |
| SDF | Download SDF |
| Canonical SMILES | ClC1=CN=C(N=C1C2=CNC3=CC=CC=C23)NC4=CC=CC(NC(C5=CC=C(NC(/C=C/CN(C)C)=O)C=C5)=O)=C4 |
| Shipping Condition | Small Molecules with Blue Ice, Modified Nucleotides with Dry Ice. |
| General tips | We do not recommend long-term storage for the solution, please use it up soon. |
| Cell experiment: [1] | |
|
Cell lines |
Jurkat and Loucy T-ALL cell lines |
|
Preparation method |
The solubility of this compound in DMSO is >10 mM. General tips for obtaining a higher concentration: Please warm the tube at 37°C for 10 minutes and/or shake it in the ultrasonic bath for a while.Stock solution can be stored below -20°C for several months. |
|
Reaction Conditions |
72 hours IC50: 50 nM (for Jurkat cells), 0.55 nM (for Loucy T-ALL cells) |
|
Applications |
As a CDK7 inhibitor, THZ1 potently inhibited proliferation of Jurkat and Loucy T-ALL cell lines with IC50 values of 50 nM and 0.55 nM, respectively. |
| Animal experiment: [1] | |
|
Animal models |
Mice bearing KOPTK1 xenografts |
|
Dosage form |
10 mg/kg, twice daily for 29 days |
|
Applications |
THZ1 exhibited efficacy in a bioluminescent xenografted mouse model using the human T-ALL cell-line KOPTK1 when dosed twice daily at 10 mg/kg. THZ1waswell tolerated at these doseswith no observable body weight loss or behavioural changes, suggesting that it caused no overt toxicity in the animals. |
|
Other notes |
Please test the solubility of all compounds indoor, and the actual solubility may slightly differ with the theoretical value. This is caused by an experimental system error and it is normal. |
|
References: [1] Kwiatkowski N, Zhang T, Rahl P B, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature, 2014, 511(7511): 616-620. |
|
| Description | THZ1 is an irreversible, potent and selective inhibitor of CDK7(cyclin-dependent kinase 7) with an IC50 value of 3.2 nM. | |||||
| Targets | CDK7 | |||||
| IC50 | 3.2 nM | |||||
Quality Control & MSDS
- View current batch:
Chemical structure

Related Biological Data
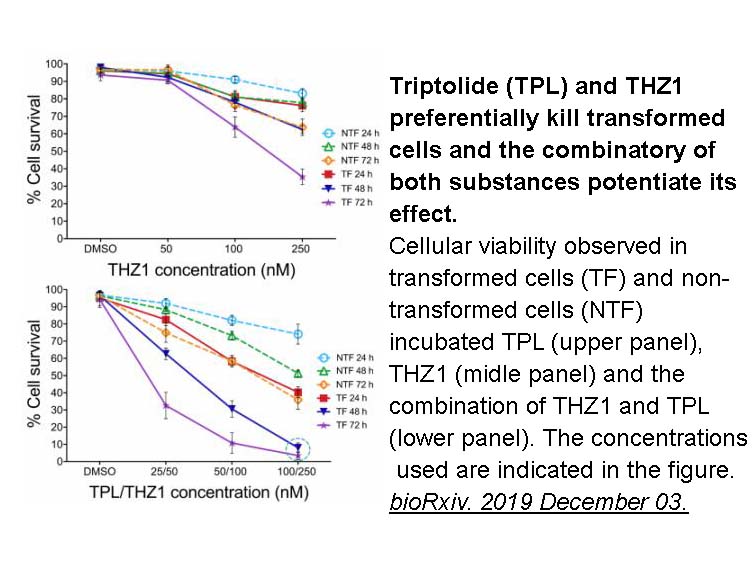
Related Biological Data
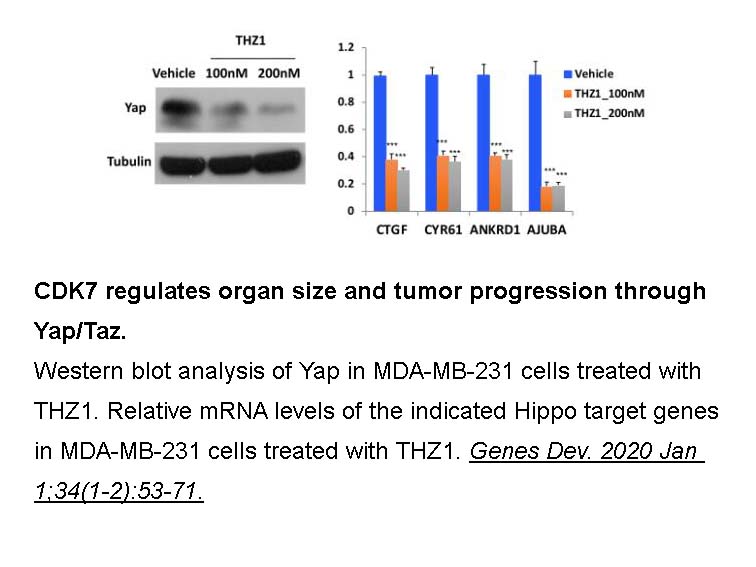
Related Biological Data
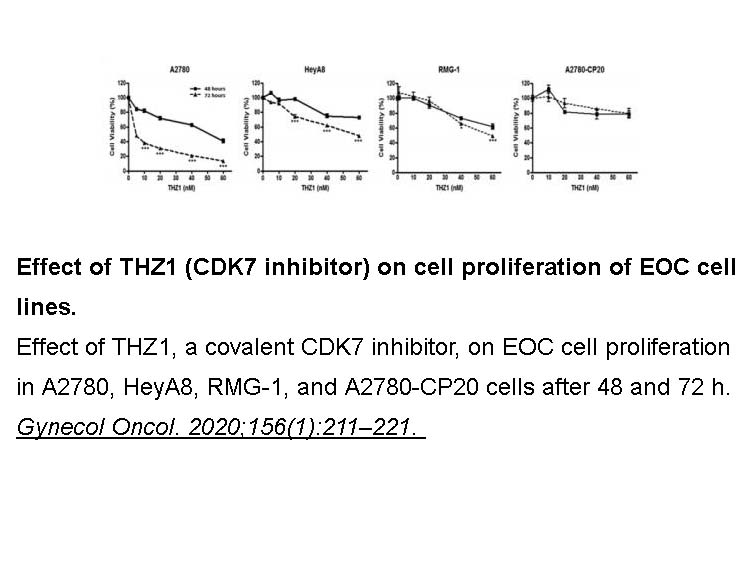
Related Biological Data
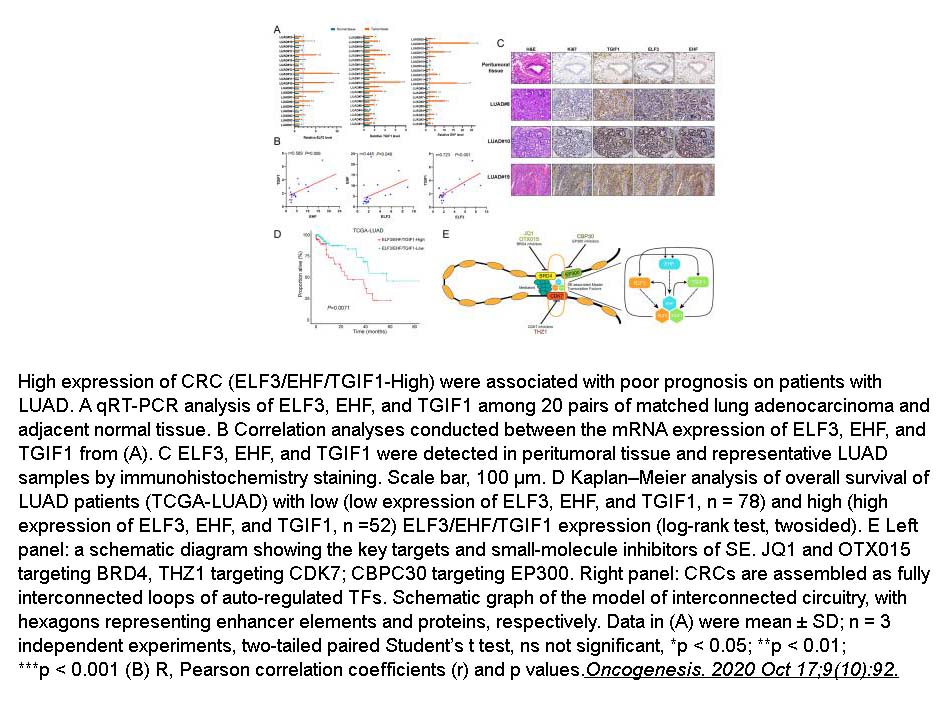
Related Biological Data
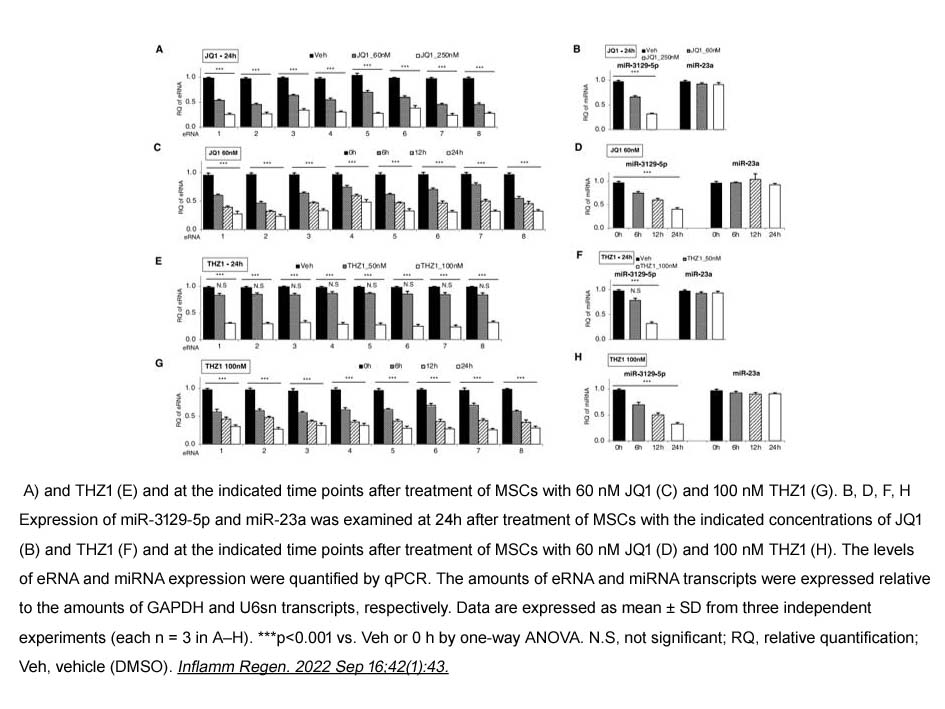
Related Biological Data
Related Biological Data