Metatranscriptomics
How to Request a Quote

Form
Please download and fill in the

Email
And then send the form to [email protected]

Feedback
We will contact you by email as soon as possible, please make sure your Email address is correct and check your email in time.
1. Overview
Metatranscriptome refers to the total content of gene transcripts in a community and varies with time and environmental changes. The techniques applied to obtain the whole expression profile in a community and to follow the dynamics of gene expression patterns over time or environmental parameters are known as metatranscriptomics. It improves our understanding of the structure, function, and adaptive mechanisms in complex communities and, together with other “omics,” is applied to fields such as medicine, biotechnology, and, particularly, in microbial ecology. Compared with the metagenomics, metatranscriptome sequencing can give us better understanding of complex microbial communities from the transcription level.APExBIO uses advanced sequencing platforms, which have high-throughput coverage that can comprehensively detect microbial information in the environment, We will help you explore the microbial diversity and dynamic changes in the sample.
3. Applications
3.1 Tibet plateau probiotic mitigates chromate toxicity in mice by alleviating oxidative stress in gut microbiota
Abstract
Heavy metal contamination in food endangers human health. Probiotics can protect animals and human against heavy metals, but the detoxification mechanism has not been fully clarified. Here, mice were supplemented with Pediococcus acidilactici strain BT36 isolated from Tibetan plateau yogurt, with strong antioxidant activity but no chromate reduction ability for 20 days to ensure gut colonization. Strain BT36 decreased chromate accumulation, reduced oxidative stress, and attenuated histological damage in the liver of mice. 16S rRNA and metatranscriptome sequencing analysis of fecal microbiota showed that BT36 reversed Cr(VI)-induced changes in gut microbial composition and metabolic activity. Specifically, BT36 recovered the expressions of 788 genes, including 34 inherent Cr remediation-relevant genes. Functional analysis of 10 unannotated genes regulated by BT36 suggested the existence of a new Cr(VI)-reduction gene in the gut microbiota. Thus, BT36 can modulate the gut microbiota in response to Cr(VI) induced oxidative stress and protect against Cr toxicity.
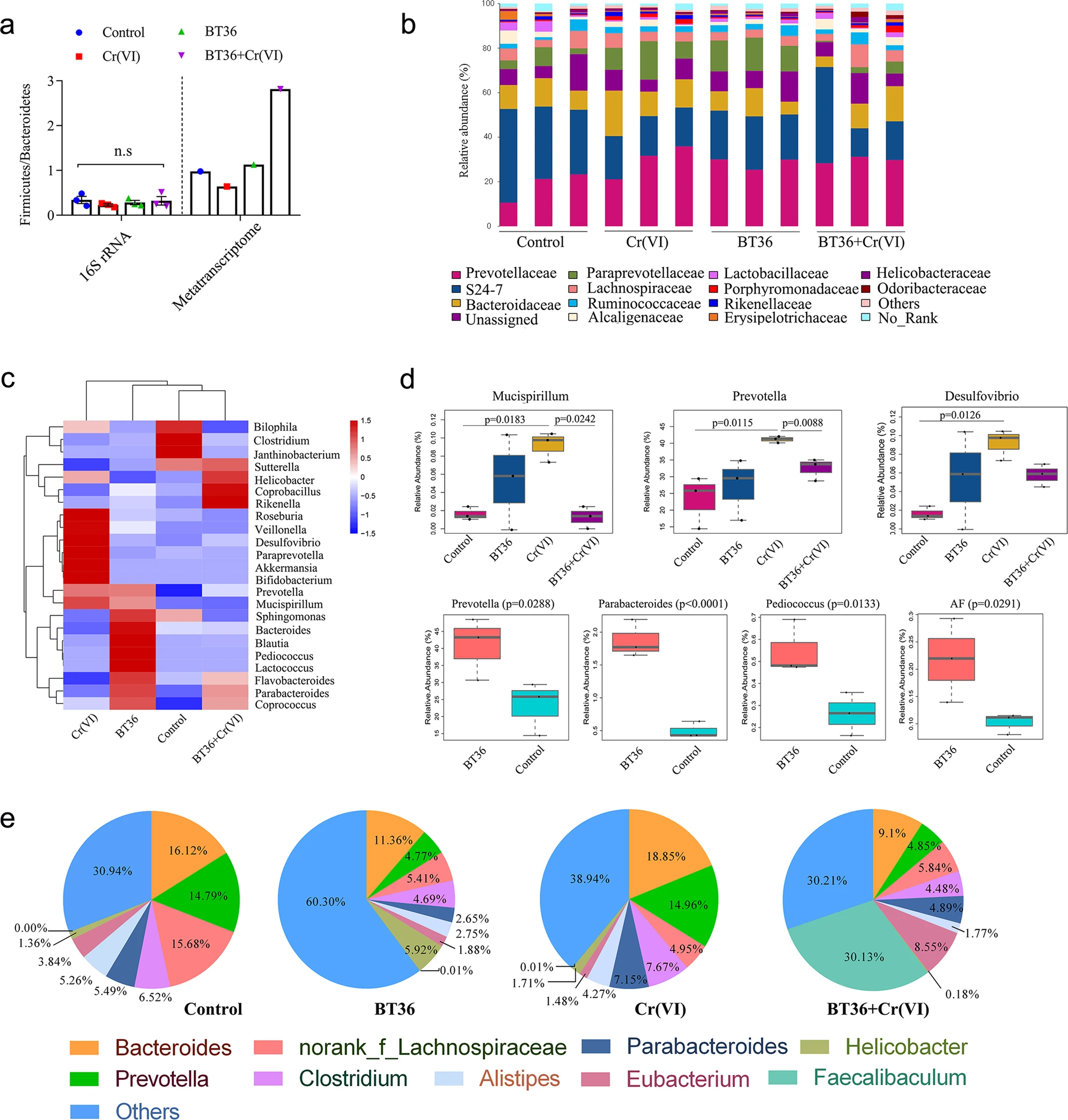
Figure 3.1.1 a The ratio of Firmicutes and Bacteroidetes. For 16S rRNA analysis, statistical analyses were conducted using the one-way ANOVA followed by Tukey’s post hoc test (n = 3). b Relative abundance (% of total reads) of bacterial 16S rRNA gene at the family level (n = 3). c Heatmap of the genus based on 16S rRNA sequencing (n = 3). The color of each rectangle in the heatmap reprensents the normalized and log-transformed relative abundance of each genus in four groups by a gradient of color from blue (low abundance, −1.5) to red (high abundance, 1.5). d Alterations in genus with statistically significant differences based on 16S rRNA sequencing (n = 3). The box- plots show the maximum, minimum and median, and the whisker indicates total range. e Taxonomic assignment of mRNA in each sample at the genus level. By default, all species whose expression abundance was less than 1% in all samples were combined with others. For d, STAMP v2.0.0 software (p < 0.05) was employed to analyze statistical significance between the fecal microbiome of four groups using ANOVA with Benjamini-Hochberg FDR multiple test correction, and to analyze statistical significance between the fecal microbiome of two groups using one sided t-test.
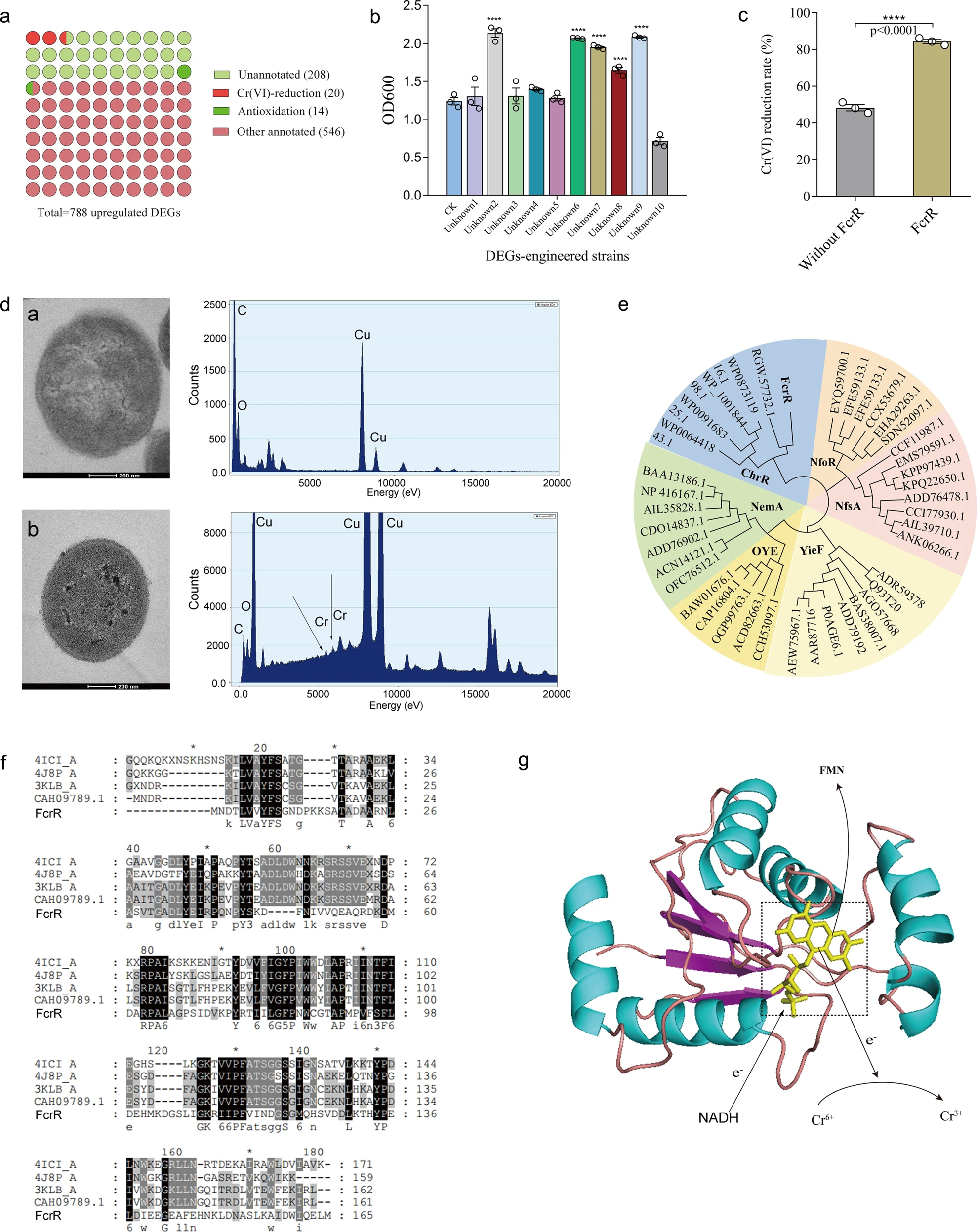
Figure 3.1.2 a The distribution of Cr remediation relevant genes in total DEGs. b The growth of engineered E. coli strains harboring DEGs in LB medium with 1 mM Cr(VI) (n = 3). CK represents E. coli BL21 containing the empty plasmid pET28a (+), unknown 1–10 represents E. coli BL21 with pET28a harboring gene comp53346, 53095, 24337, 53087, 53032, 51796, 52516, 53047, 53431, and 53299, respectively. c Cr(VI) reduction rate of engineered E. coli BL21 with 53431 (n = 3). d TEM and EDX analyses of E. coli FcrR after inoculated in LB medium and incubated overnight without Cr(VI) or with 1 mM Cr(VI), and the corresponding EDX spectra. Arrows show intracellular Cr(III) precipitates, and scale bars: 200 nm. e Evolutionary relationship of FcrR with known chromate reductases from different genera. A composite evolutionary tree with the FcrR was generated using the Neighbor-Joining method based on the reported chromate reductase families (NfoR, NfsA, YieF, OYE, NemA, and ChrR). f Sequence analysis of FcrR homologs. The highly conserved residues are highlighted in black, and other residues with high levels of similarity are highlighted in gray. g Structural modeling of FcrR. For b, data significance was analyzed using the one-way ANOVA followed by Tukey’s post hoc test. For c, data significance was analyzed using student’s t-test. Significance was marked as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
4. References
[1] Feng, P., Ye, Z., Han, H. et al. Tibet plateau probiotic mitigates chromate toxicity in mice by alleviating oxidative stress in gut microbiota. Commun Biol 3, 242 (2020).


