Proteinase K
Proteinase K is a broad-spectrum serine protease. Our product is derived from recombinant Pichia pastoris strains expressing the endoproteinase gene originally sourced from the fungus Tritirachium album limber. It is a highly active enzyme widely used in the hydrolysis of a variety of proteins and enzymatic contaminants (including endonucleases, exonucleases, DNases, and RNases). Proteinase K is therefore commonly used for DNA preparations as it does not impair DNA integrity. Proteinase K preferentially cleaves peptide bonds adjacent to the carboxyl end of hydrophobic amino acids (aliphatic, aromatic, etc.) and exhibits excellent performance under various conditions such as different pH values, buffer types, detergents (e.g. SDS), chelating agents (e.g. EDTA), and temperatures. Proteinase K is also used for enzyme mapping and the removal of unwanted enzymes from DNA preparations to enhance cloning efficiency. Except the isolation of genome, it can also take a job in detection of enzyme localization or removal of enzymes from DNA to improve cloning efficiency.
Appropriate working concentration of proteinase K is always among the range of 0.05 to 1 mg/mL. The activity of the enzyme can be stimulated by 0.2 to 1% SDS or by 1 to 4 M urea. It is activated by calcium (1-5 mM). Calcium ions do not affect the enzyme activity, but it contributes to the thermal stability and protects the proteinase from autolysis. Calcium ion has a regulatory function for the substrate binding site of proteinase K. The enzyme is inactivated by DIFP or PMSF. However, it is not inhibited by EDTA, iodoacetic acid, trypsin-specific inhibitor TLCK, chymotrypsin-specific inhibitor TPCK, and p-chloromercuribenzoate.
We recommend an optimum pH of 7.5 to 8.0 and optimum temperature at 50 to 55°C. Rapid denaturation will occur above 65°C. It is inactivated under 95°C for 10 min.
References:
[1] Kraus, E., et al. Proteinase K from the Mold Tritirachium album limber, Specificity and Mode of Action. Z. Physiol. Chem., 357:939; 1976.
[2] Jany, K.D., et al. Amino Acid Sequence of Proteinase K from the Mold, Tritirachium albumlimber. Proteinase K; a Subtilisin-related Enzyme with Disulfide Bonds. FEBS Letter, 199,139.1986.
| CAS No. | 39450-01-6 | SDF | |
| Concentration | > 600 U/mL (approximately 20 mg/mL) | ||
| Formula | M.Wt | 29.3 kDa | |
| Solubility | Soluble in 20 mM Tris-HCl, 1 mM CaCl2, 50% Glycerol, pH 7.4 | Storage | Store at -20°C |
| Stability | pH range: 4.0 to 12.5, temperature range: 25°C to 65°C. | ||
| General Tips | APExBIO guarantees optimal performance of this product for 18 months after date of delivery under the appropriate temperature and condition. | ||
| Shipping Condition | Ship with blue ice | ||
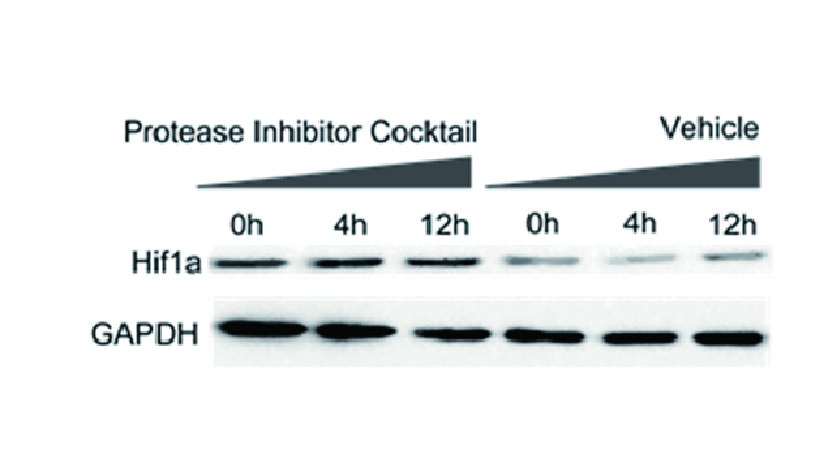
- 1. Yu Wei, Yujie Zhou, et al. "Candida albicans Extracellular Vesicles Upregulate Nrg1 Transcription Repressor to Inhibit Self-Hyphal Development and Candidemia." Int J Mol Sci. 2026 Jan 3;27(1):495. PMID: 41516377
- 2. Jack J. Devlin, Cleverson Lima, Yuta Kawarasaki. "Prevalence and consequences of microplastic ingestion in the world's southernmost insect, Belgica antarctica." Science of The Total Environment Volume 1004, 15 November 2025, 180800. PMID: 41172662
- 3. Nicole DeFoor, Swagatika Paul, et al. "Remdesivir increases mtDNA copy number causing mild alterations to oxidative phosphorylation." Sci Rep. 2023 Sep 15;13(1):15339. PMID: 37714940
- 4. Huang J, Chen P, et al. "Multi‐Omics Analysis Reveals Translational Landscapes and Regulations in Mouse and Human Oocyte Aging." Advanced Science, 2023: 2301538. PMID: 37401155
- 5. Hira Khan, Takashi Ochi, et al. "Plant PAXX has an XLF‐like function and stimulates DNA end‐joining by the Ku‐DNA ligase IV‐XRCC4 complex." Plant J. 2023 Oct;116(1):58-68. PMID: 37340932
- 6. Penghui Zhang, Xizhe Liu, et al. "Effect of cyclic mechanical loading on immunoinflammatory microenvironment in biofabricating hydroxyapatite scaffold for bone regeneration." Bioact Mater. 2021 Mar 9;6(10):3097-3108. PMID: 33778191
Quality Control & DataSheet
- View current batch:
-
Enzyme activity: ≥ 30 units/mg of protein
- COA (Certificate Of Analysis)
- Datasheet
Specific activity: ≥ 40 units/mg of protein
Purity: ≥ 95%(Native-PAGE)
RNase and DNase free