I-BET-762
I-BET-762 is a highly potent inhibitor of BET with IC50 values of 32.5–42.5 nM [1].
I-BET-762 is found to have high affinity with BET (Kd of 50.5–61.3 nM). It binds to the acetyl-lysine (AcK)-binding pocket of BET and competes with AcK. The structure of I-BET-762 allows it to bind with BET in ratio of 2:1 results in the high affinity of it. I-BET-762 also has high selectivity, it has no interaction with other bromodomain-containing proteins. I-BET-762 is reported to downregulate the expression of the genes which are induced by LPS, thus causing the decreased expression of LPS-inducible cytokines and chemokines. In vivo assay shows that I-BET-762 has the anti-inflammatory potential. Treatment of I-BET-762 can cure the mice which have started to develop symptoms of inflammatory disease [1].
References:
[1] Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, White J, Kirilovsky J, Rice CM, Lora JM, Prinjha RK, Lee K, Tarakhovsky A. Suppression of inflammation by a synthetic histone mimic. Nature. 2010 Dec 23;468 (7327):1119-23.
- 1. Liuting Chen, Lu Wang, et al. "Identification and genetic validation of leukemia inhibitory factor super‐enhancers in acute respiratory distress syndrome and lung cancer." Cell Biochem Funct. 2024 Jun;42(4):e4031. PMID: 38760985
- 2. Chenyang Fan, Xiaohong Guo, et al. "BRD4 inhibitors broadly promote erastin-induced ferroptosis in different cell lines by targeting ROS and FSP1." Discov Oncol. 2024 Apr 3;15(1):98. PMID: 38565708
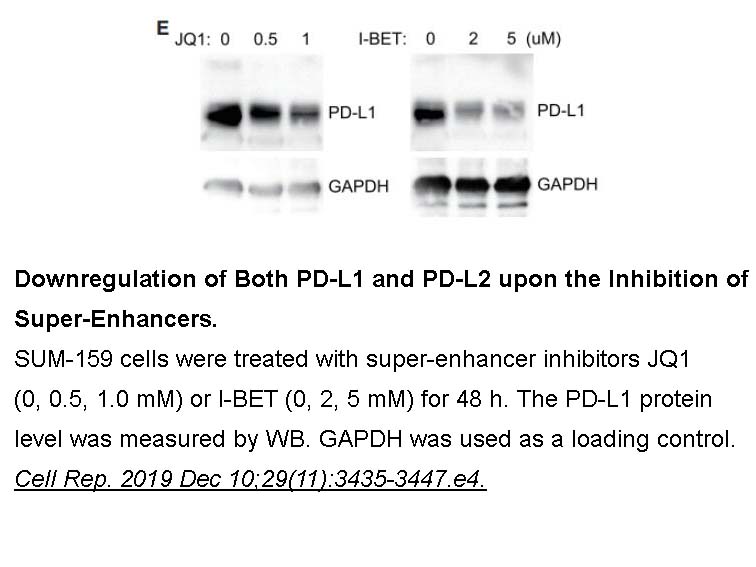
- 3. Xu Y, Wu Y, et al. "A Tumor-Specific Super-Enhancer Drives Immune Evasion by Guiding Synchronous Expression of PD-L1 and PD-L2." Cell Rep. 2019 Dec 10;29(11):3435-3447.e4. PMID:31825827
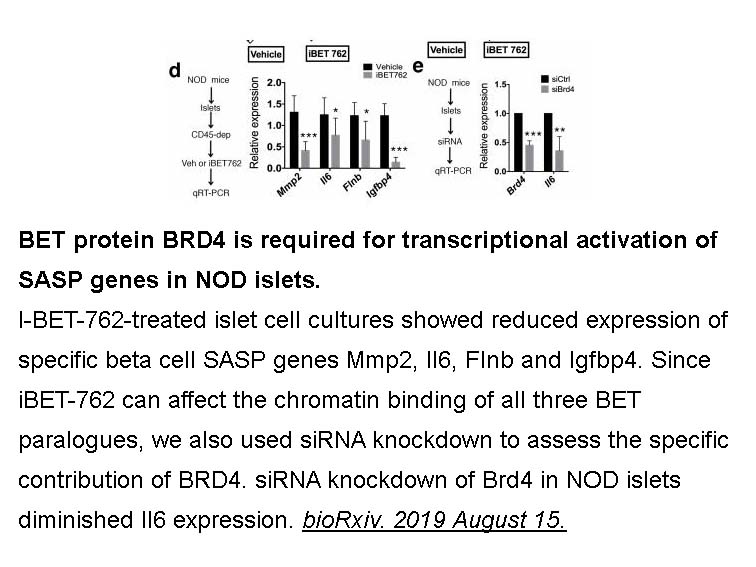
- 4. Peter J. Thompson, Ajit Shah, et al. "BET proteins are required for transcriptional activation of the senescent beta cell secretome in Type 1 Diabetes." bioRxiv. 2019 August 15.
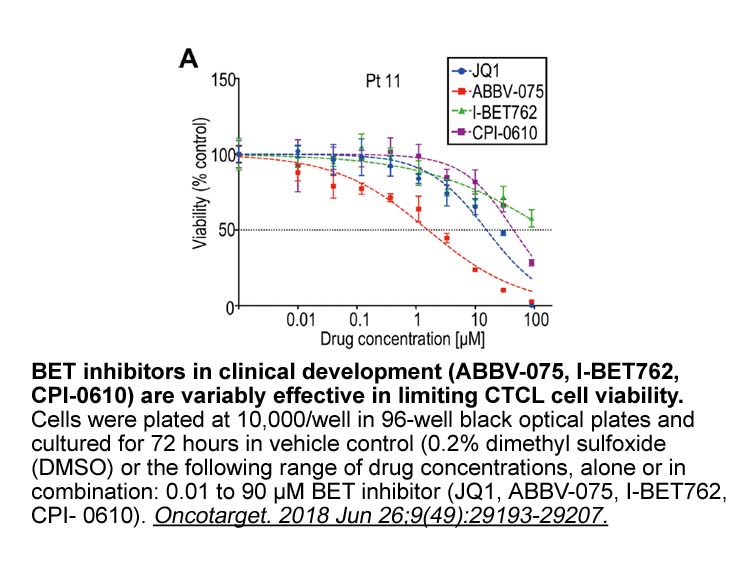
- 5. Kim SR, Lewis JM, et al. "BET inhibition in advanced cutaneous T cell lymphoma is synergistically potentiated by BCL2 inhibition or HDAC inhibition." Oncotarget. 2018 Jun 26;9(49):29193-29207. PMID:30018745
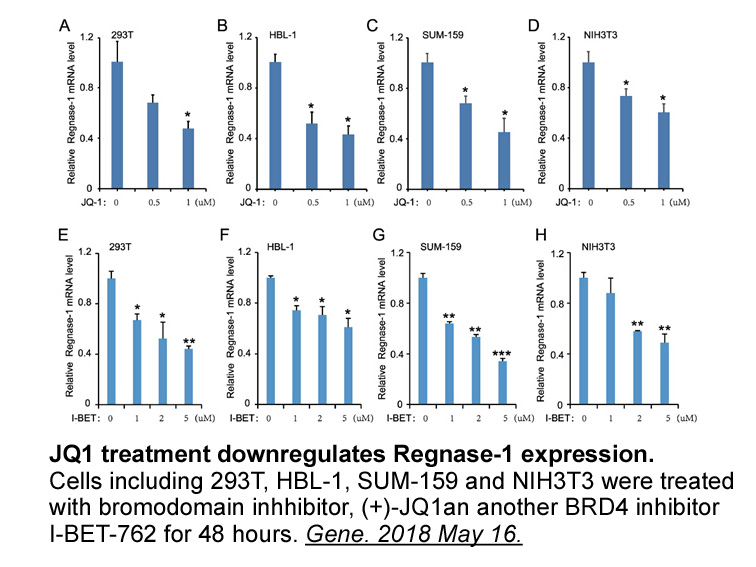
- 6. Yang R, Wu Y, et al. "A super-enhancer maintains homeostatic expression of Regnase-1." Gene. 2018 May 16. pii: S0378-1119(18)30542-0. PMID:29777912
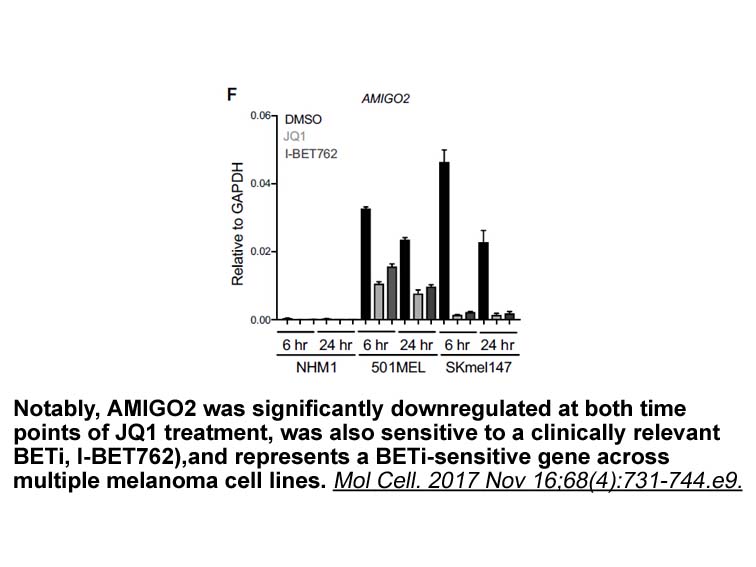
- 7. Fontanals-Cirera B, Hasson D, et al. "Harnessing BET Inhibitor Sensitivity Reveals AMIGO2 as a Melanoma Survival Gene." Mol Cell. 2017 Nov 16;68(4):731-744.e9. PMID:29149598
- 8. Ronald M. Evans,Michael Downes,et al. "Increasing storage of vitamin a, vitamin d and/or lipids." US Patent App. 15, 2016.
| Physical Appearance | A solid |
| Storage | Store at -20°C |
| M.Wt | 423.9 |
| Cas No. | 1260907-17-2 |
| Formula | C22H22ClN5O2 |
| Solubility | ≥21.19 mg/mL in DMSO; insoluble in H2O; ≥13.93 mg/mL in EtOH with ultrasonic |
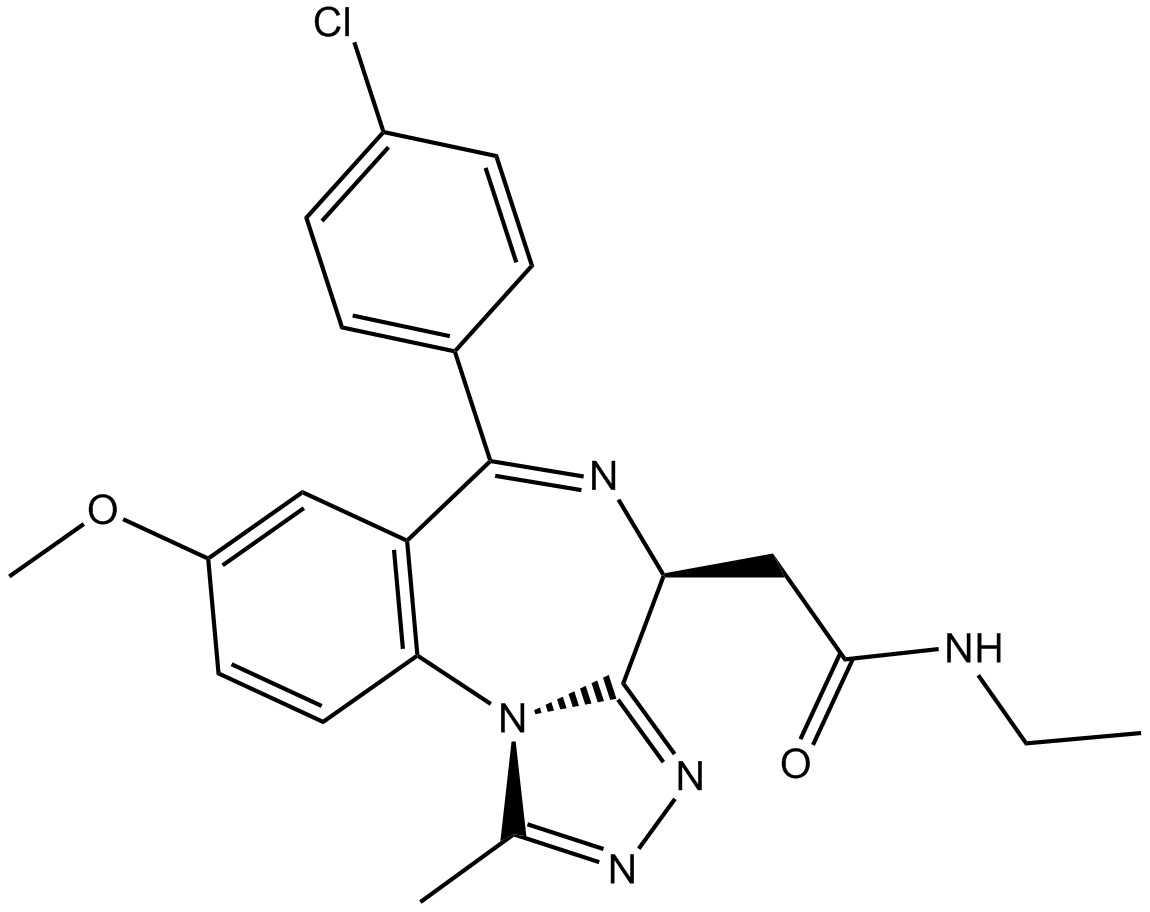
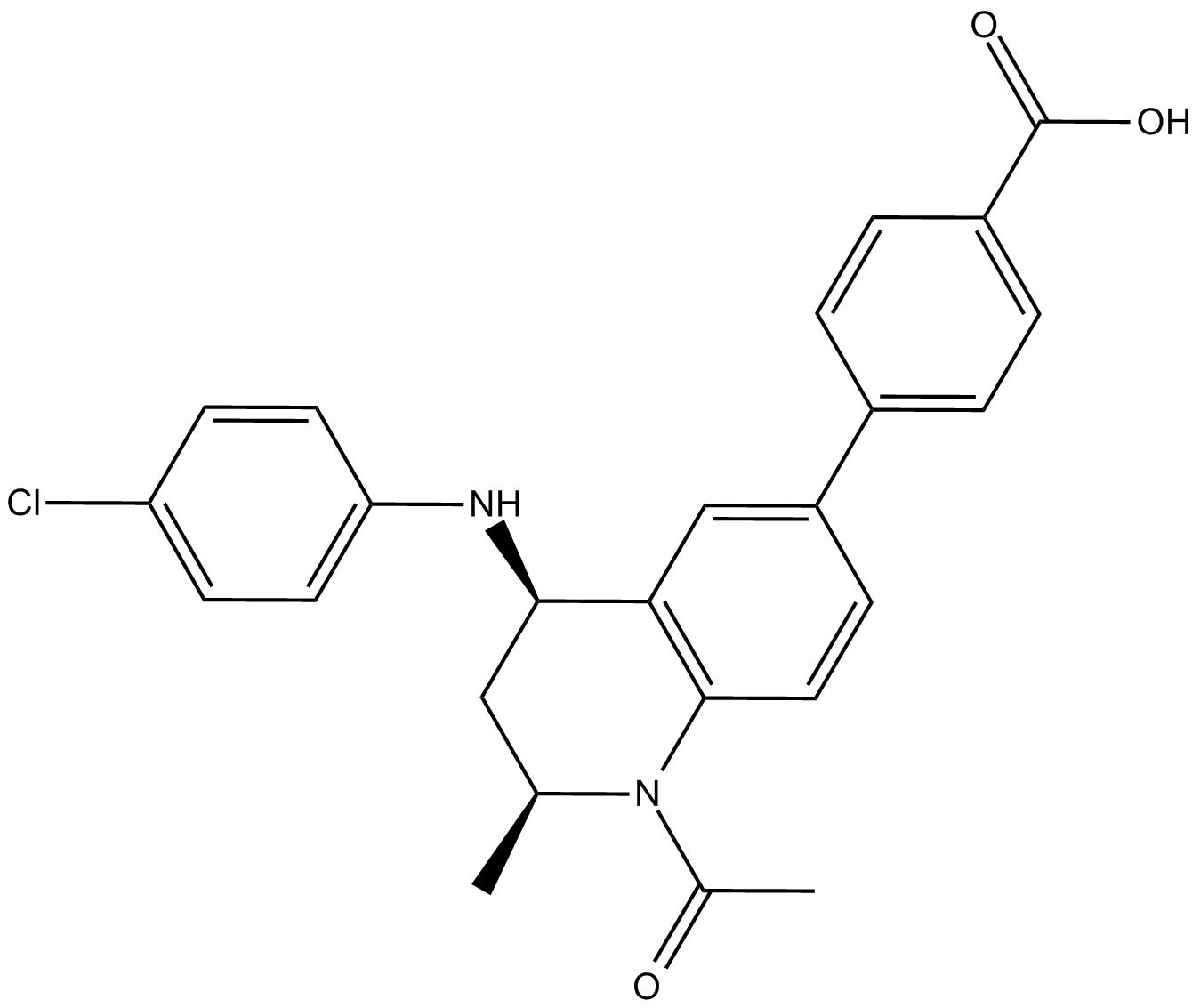
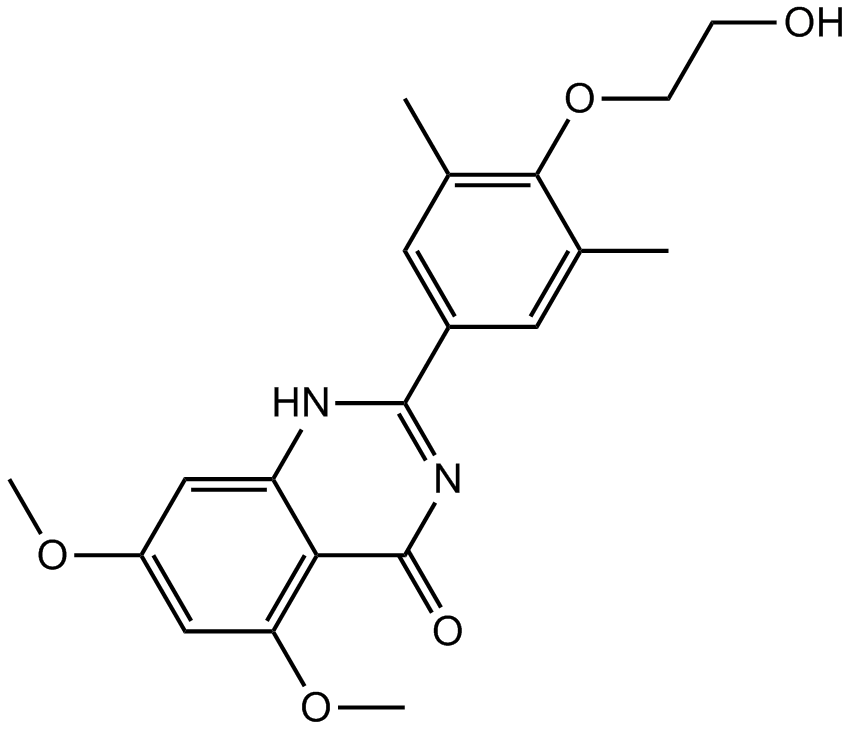
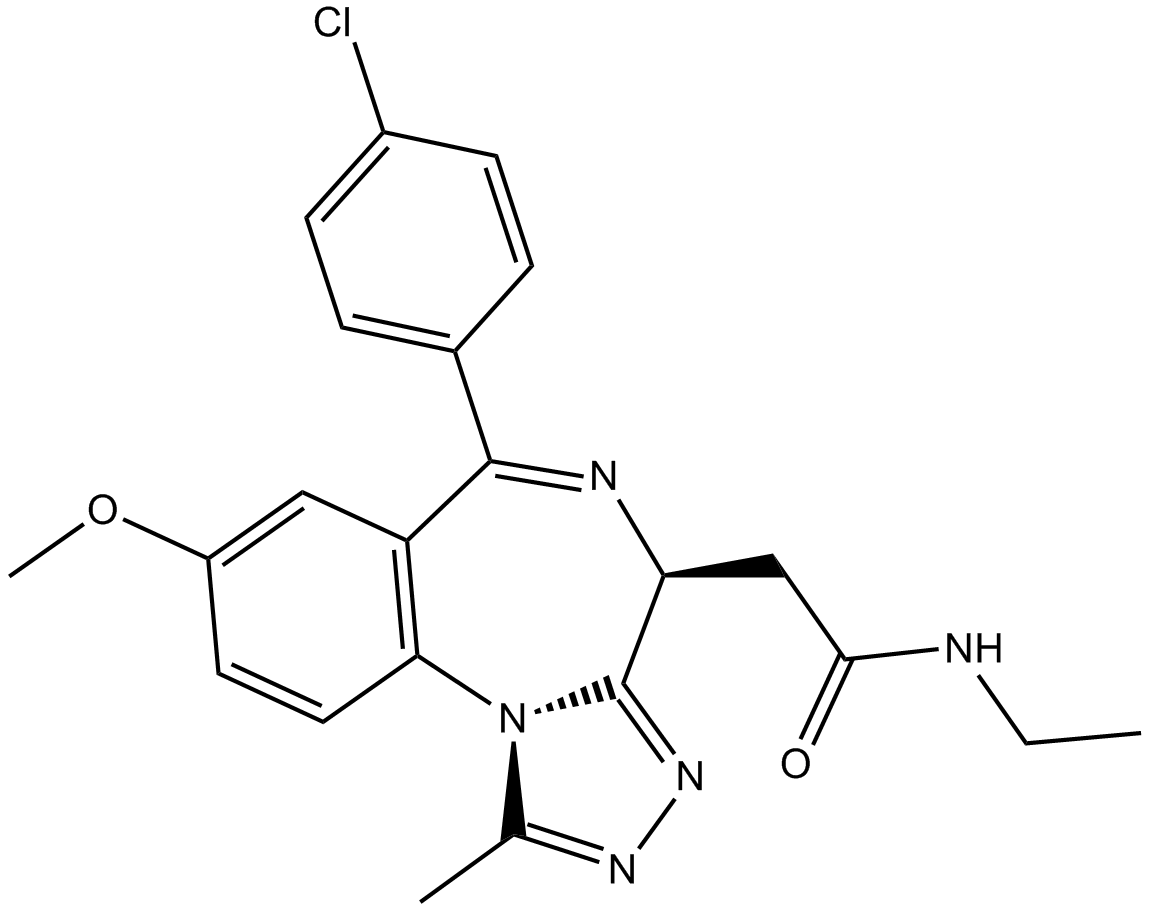
| Chemical Name | 2-[(4S)-6-(4-chlorophenyl)-8-methoxy-1-methyl-4H-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-N-ethylacetamide |
| SDF | Download SDF |
| Canonical SMILES | CCNC(C[C@@H]1N=C(c(cc2)ccc2Cl)c(cc(cc2)OC)c2-[n]2c1nnc2C)=O |
| Shipping Condition | Small Molecules with Blue Ice, Modified Nucleotides with Dry Ice. |
| General tips | We do not recommend long-term storage for the solution, please use it up soon. |
| Kinase experiment [1]: | |
|
Fluorescence resonance energy transfer (FRET) titrations |
I-BET is titrated against BRD2 (200 nM), BRD3 (100 nM) and BRD4 (50 nM) in 50 mM HEPES pH7.5, 50 mM NaCl, 0.5 mM CHAPS in the presence of tetra-acetylated Histone H4 peptide (200 nM). After equilibrating for 1 hour, the bromodomain protein : peptide interaction is detected using FRET following the addition of 2nM Europium cryptate labelled streptavidin and 10 nM XL-665-labelled anti-6His antibody in assay buffer c |
| Description | I-BET-762 is a highly potent inhibitor of bromo and extra C-terminal domain (BET) family with IC50 values of 32.5–42.5 nM. | |||||
| Targets | BET | |||||
| IC50 | 32.5–42.5 nM | |||||
Quality Control & MSDS
- View current batch:
Chemical structure
Related Biological Data
Related Biological Data
Related Biological Data
Related Biological Data
Related Biological Data