EdU Flow Cytometry Assay Kits (488)
Measuring cell proliferation and cell cycle are a fundamental method to assess cell health, determine genotoxicity, and evaluate drug’s pharmacodynamic effect. The common method is measuring DNA synthesis directly. In previous experiments, there are several approaches such as the incorporation of radioactive nucleosides (3H-thymidine) or BrdU. Here, we introduce one new method, click chemistry - CuAAC (Copper-Catalyzed Azide-Alkyne Cycloaddition), and the use of this reaction in direct measurement of S-phase DNA synthesis in cell cycle. "Click Chemistry" is a term that was introduced by K. B. Sharpless in 2001, it’s a set of biocompatible small molecule reactions commonly used in bioconjugation, allowing the joining of substrates with specific biomolecules (such as a reporter molecule).
A nucleoside analog of thymidine, EdU (5-ethynyl-2’-deoxyuridine), can be incorporated into DNA strand during DNA synthesis. The alkynyl group of EdU is a biologically inert group that will undergo an extremely selective reaction with dye’s azido via a CuAAC reaction to afford an 1,2,3-triazole product. EdU and 6-FAM Azide possess biologically unique moieties to fluorescently label DNA of proliferating cells, producing low backgrounds and high detection sensitivities. This CuAAC reaction affords superior regioselectivity and quantitative transformation under extremely mild conditions. The reaction is a highly efficient and can be used in a quantitative manner via Flow Cytometry, fluorimeter or fluorescence microscope.
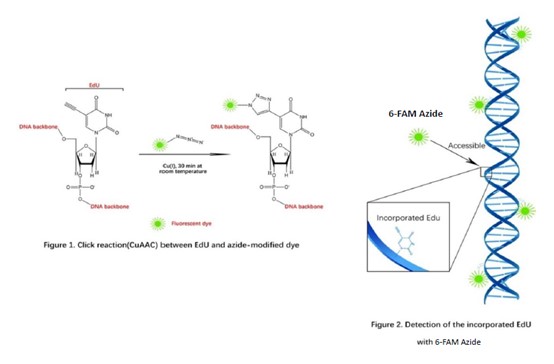
EdU is an ideal alternative of BrdU since it’s independent of harsh DNA denaturing conditions (typically using HCl, heat, or digestion with DNase). In BrdU-based assays, it requires extra DNA denaturation to expose the BrdU to an anti-BrdU antibody which is huge compared to fluorescent dye used in EdU-based assays. Unlike bulky antibodies, alkyne and azide groups are very small in size, therefore 6-FAM Azide gain access to the DNA under standard aldehyde-based fixation and detergent permeabilization. Samples after BrdU-based assay may possess signal alteration of the cell cycle distribution because of the destruction of antigen recognition sites when using the acid denaturation method. In contrast, EdU-based assay is compatible with cell cycle dyes. The EdU assay can also be multiplexed with antibodies against surface and intracellular markers. However, some reagents or antibody conjugates may not be compatible with the EdU detection reaction and may need some additional steps to ensure compatibility.
6-FAM Azide (Excitation maximum: 496 nm Emission maximum: 516 nm)
- 1. Yun Gao, Xiaobin Fang, et al. "Protocol for generating human CAR-engineered macrophages by Vpx-containing lentivirus." STAR Protoc. 2024 Sep 28;5(4):103350 PMID: 39342619
- 2. Zhen-Zhu Zhao, Xu-Bo Liang, et al. "Diterpenoids with Potent Anti-Psoriasis Activity from Euphorbia helioscopia L." Molecules. 2024 Aug 29;29(17):4104 PMID: 39274957
- 3. Shuhan Liu, Shijie Lv, et al. "The signature genes of cuproptosis associates with tumor immune microenvironment and predicts prognosis in kidney renal clear cell carcinoma." Front Oncol. 2024 Aug 14:14:1409620. PMID: 39206152
Simple works in less time with no denaturation steps or harsh treatment
High efficiency with robust brightness
Compatible with some antibodies and cell cycle dyes
High consistency because of independence of variable anti-BrdU antibody
| Components | K1176-100 Test |
| EdU (Component A) | 20 mg |
| 6-FAM Azide (Component B) | 1 vial |
| DMSO (Component C) | 8.5 mL |
| CuSO4 (100 mM Aqueous Solution) (Component D) | 1 mL |
| EdU Buffer Additive (Component E) | 400 mg |
Store the kit at -20ºC away from light and moisture, stable for 1 year. | |