Chromatin/Epigenetics
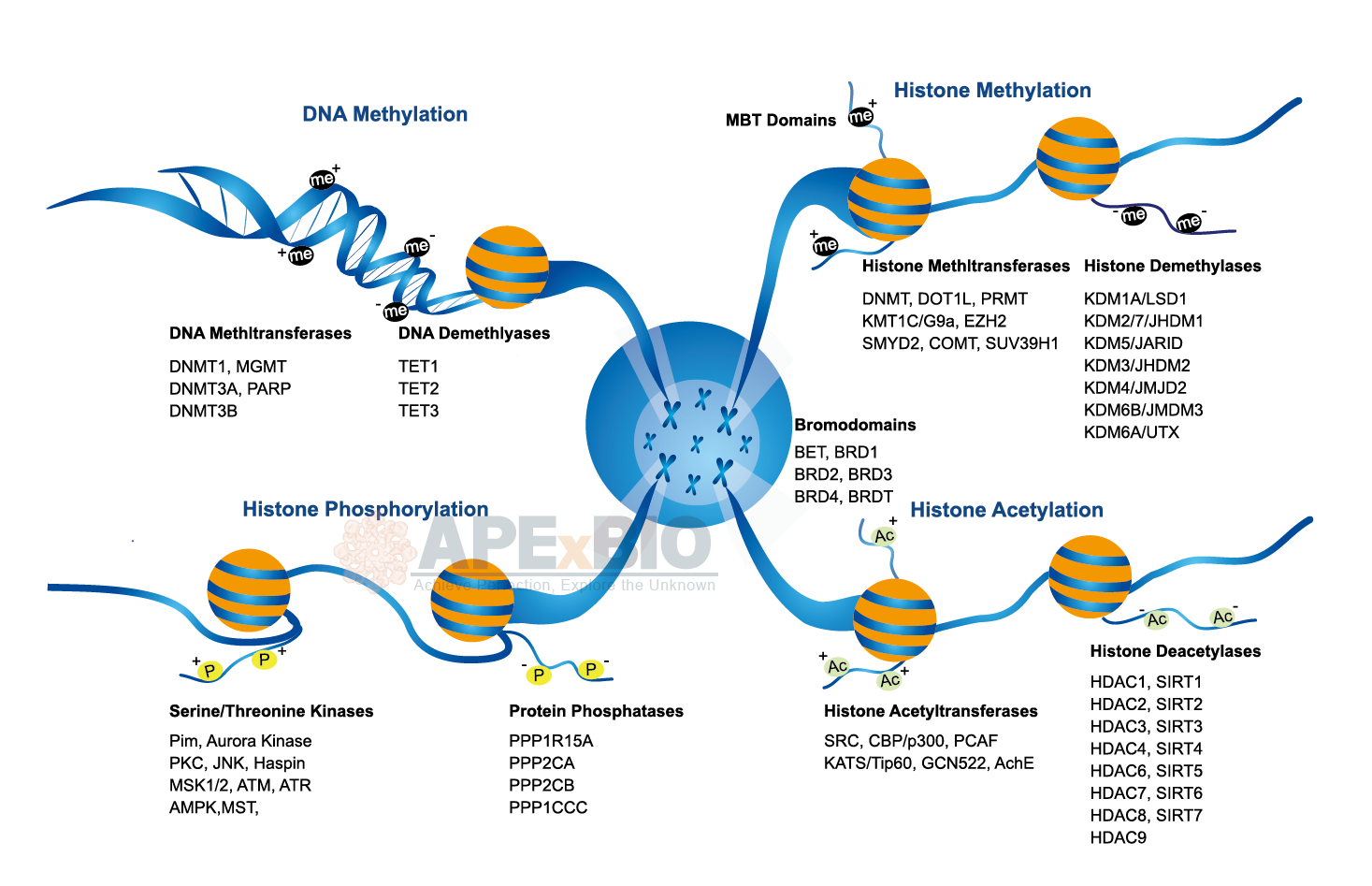
Epigenetics is the heritable modifications in gene expression that is not associated with changes in DNA sequence. Epigenetic modifications occur mostly on DNA or on the histone octamer. There are several types of epigenetics modifications, DNA methylation by DNA-methyl transferase (DNMT) and covalent modification of histones (e.g. acetylation, methylation, phosphorylation and ubiquitination). Histone acetylation by histone acetyltransferases (HATs) is involved in transcriptional activation, whereas histone deacetylation by histone deacetylases (HDACs) is connected with transcriptional repression. Histone demethylation is associated with lysine-specific demethylase (LSD) and JmjC domain containing histone demethylase (JHDM).
The nucleosome is consisted of four histone proteins (H2A, H2B, H3, and H4), they are primary building block of chromatin. The addition and removal of specific chemical groups refers to as epigenetic marks, it regulates chromatin structure and affects gene expression. Moreover, RNA is intimately involved in the formation of a repressive chromatin state.
Epigenetic mechanism responds to environmental changes at the cellular level and thus influences cellular plasticity. Chromatin and epigenetic regulation play a significant role in the programming of the genome during development and stress response, defects in epigenetics can lead to cancer, inflammation and metabolic disorders etc.
-
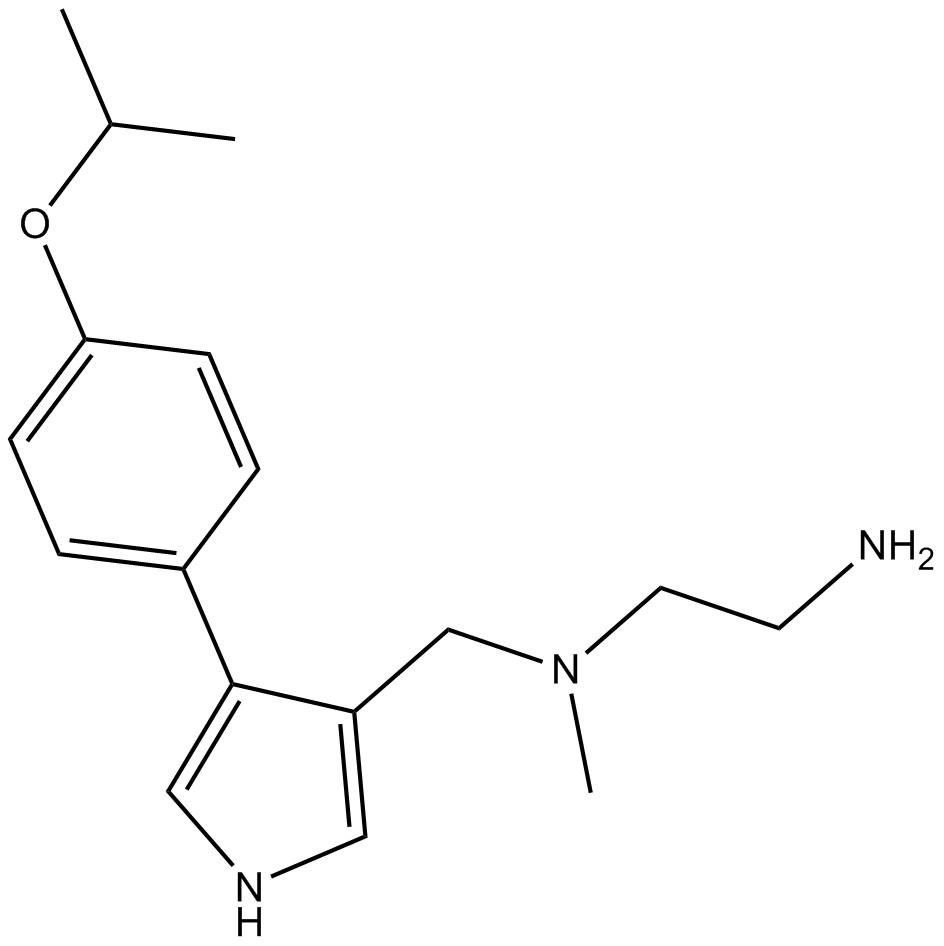 B6183 MS0231 CitationSummary: type I PRMTs inhibitor
B6183 MS0231 CitationSummary: type I PRMTs inhibitor -
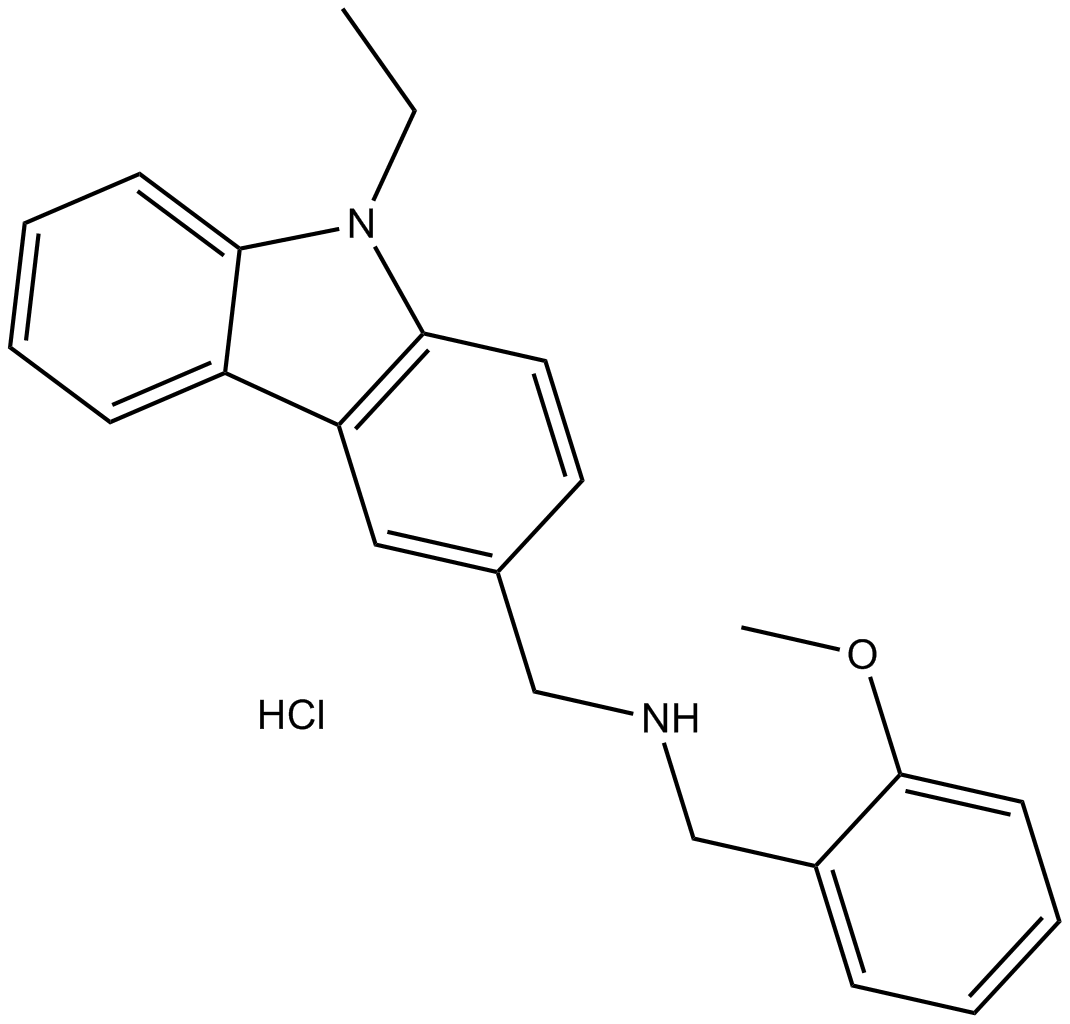 B7806 HLCL-61Summary: PRMT5 inhibitor
B7806 HLCL-61Summary: PRMT5 inhibitor -
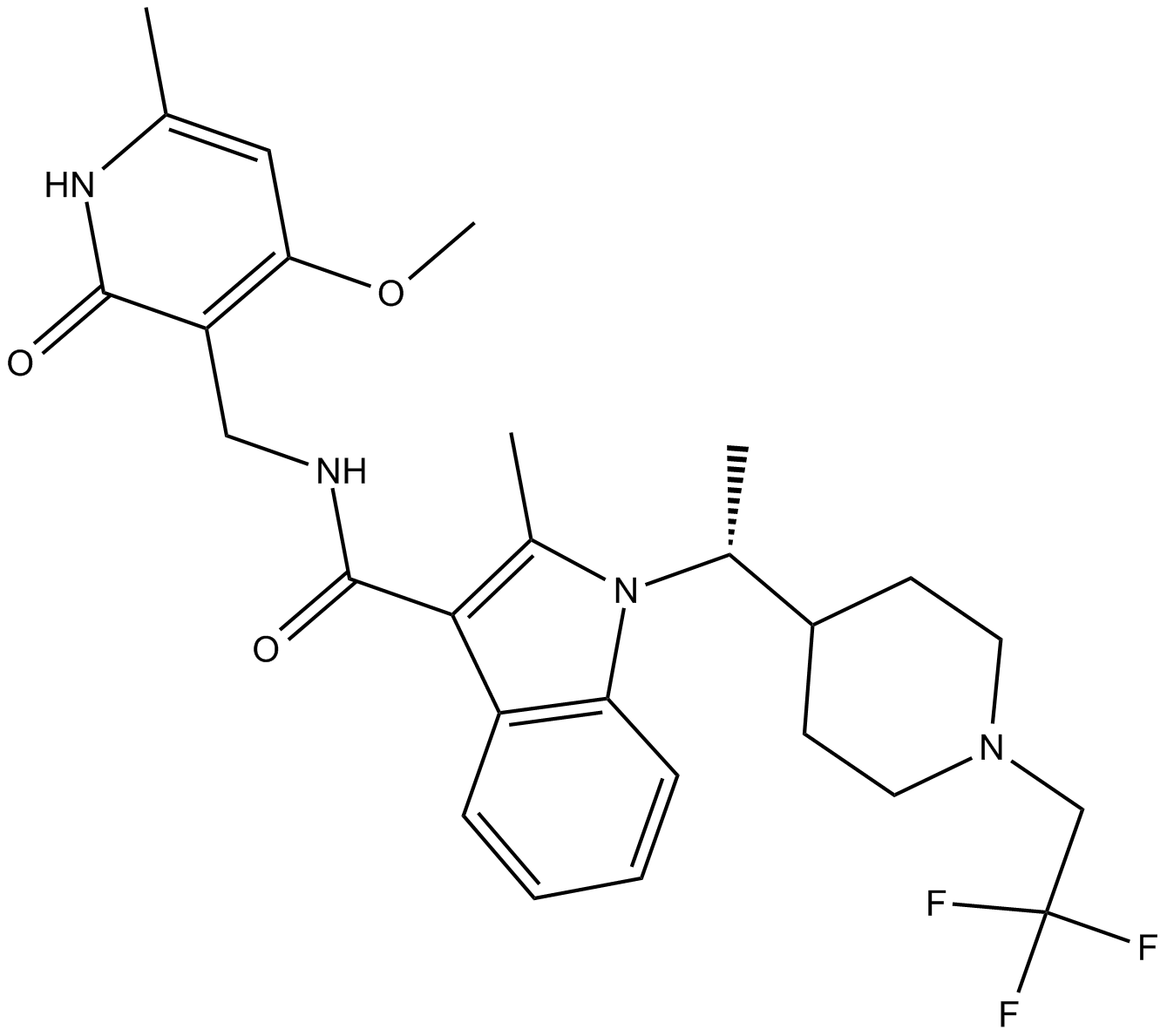 B7819 CPI-1205Summary: EZH2 inhibitor
B7819 CPI-1205Summary: EZH2 inhibitor -
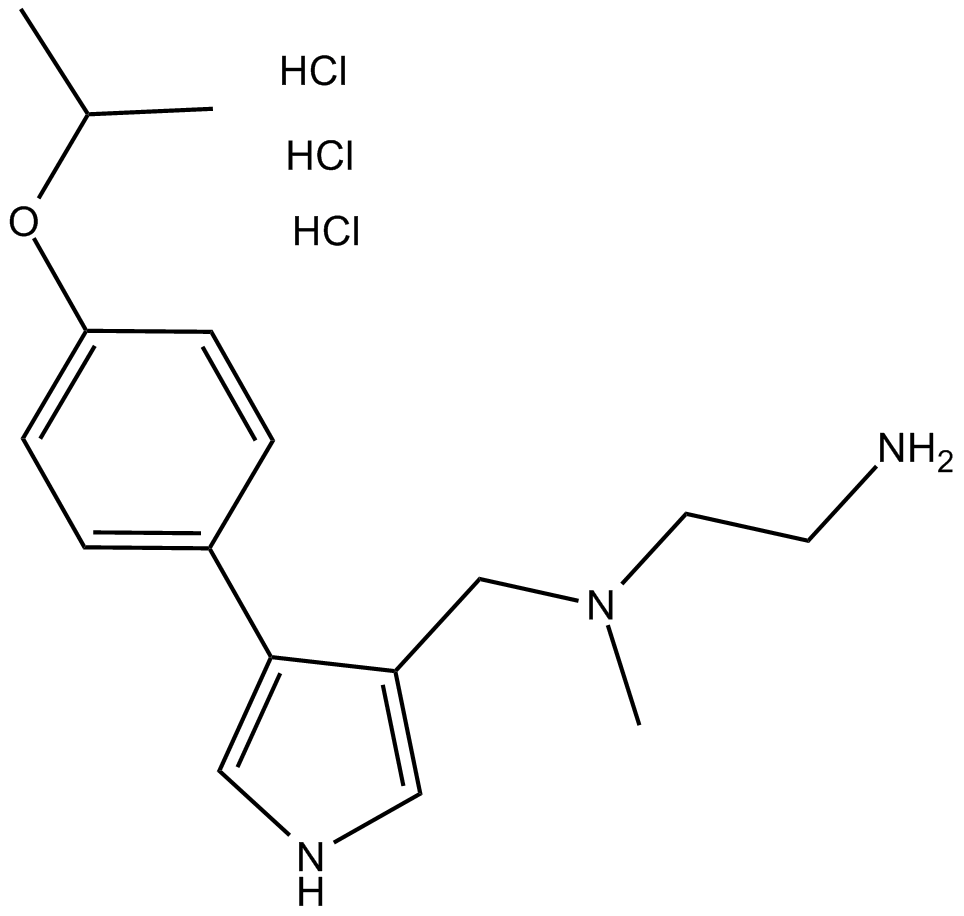 C3503 MS023 (hydrochloride)Summary: type I PRMTs inhibitor
C3503 MS023 (hydrochloride)Summary: type I PRMTs inhibitor -
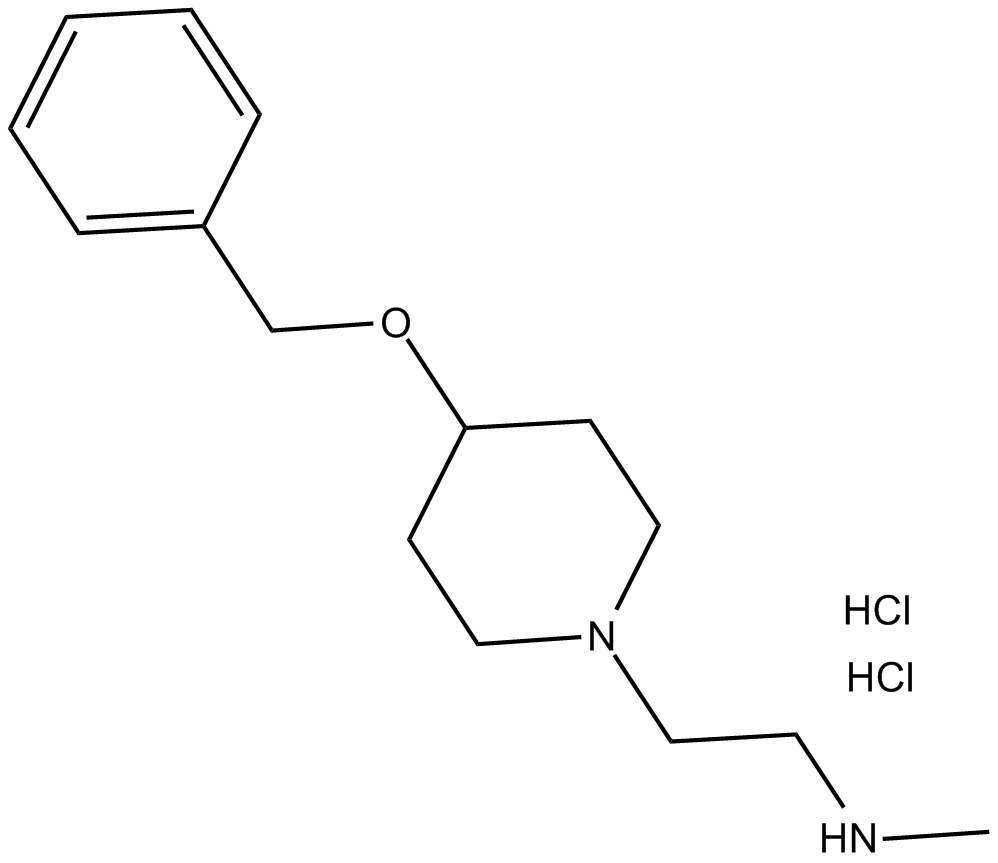 C3776 MS049 (hydrochloride)Summary: PRMT4 and PRMT6 inhibitor
C3776 MS049 (hydrochloride)Summary: PRMT4 and PRMT6 inhibitor -
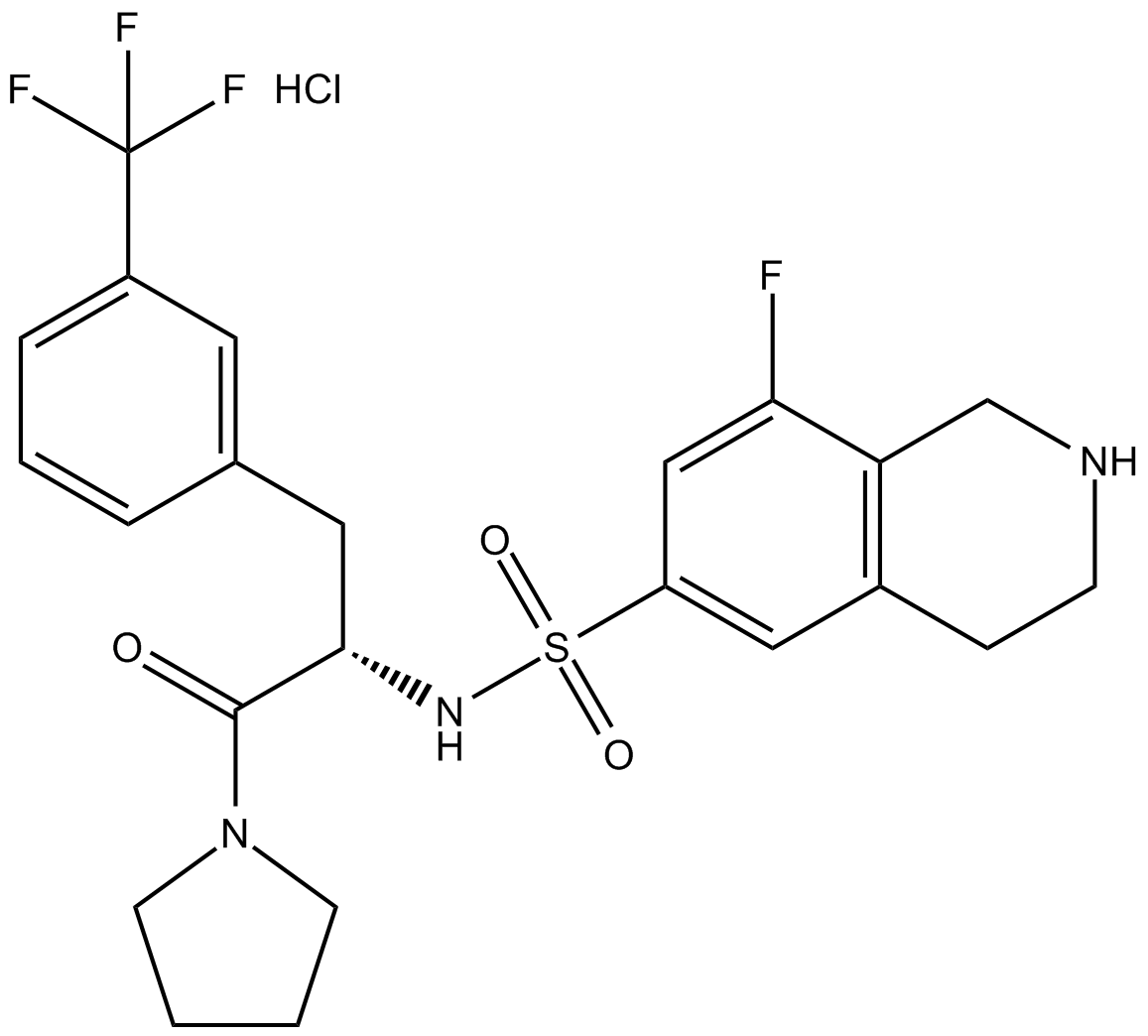 C4834 (S)-PFI-2 (hydrochloride)Summary: Negative control of (R)-PFI 2 hydrochloride
C4834 (S)-PFI-2 (hydrochloride)Summary: Negative control of (R)-PFI 2 hydrochloride -
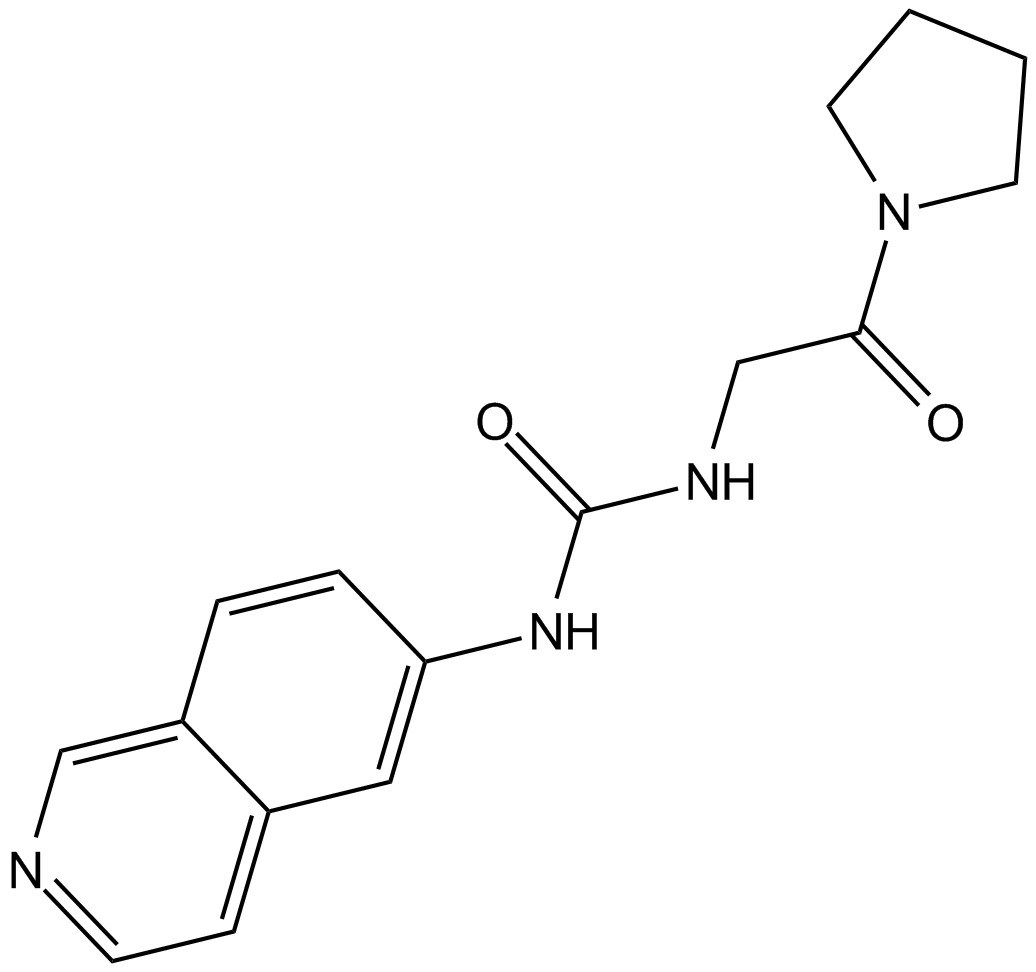 B6048 SGC707Summary: PRMT3 inhibitor
B6048 SGC707Summary: PRMT3 inhibitor

