EZ Cap™ mCherry mRNA (5mCTP, ψUTP)
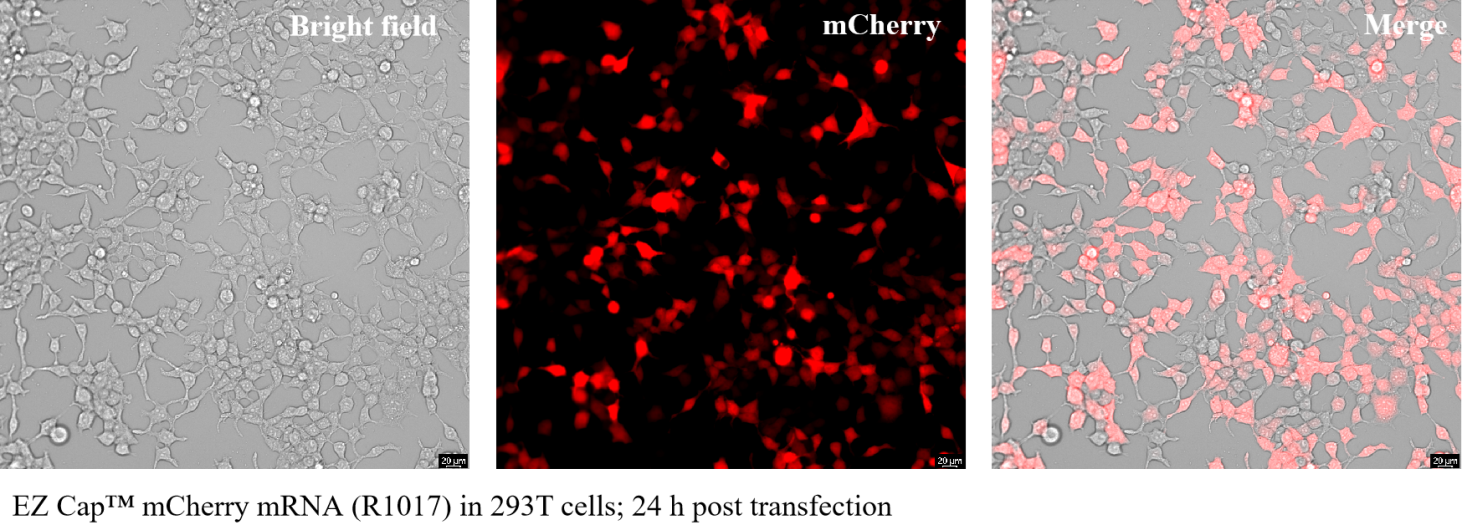
mCherry mRNA encodes the red fluorescent protein mCherry. mCherry is a derivative of the red fluorescent protein DsRed, which is isolated from the sea anemone (Discosoma). mCherry is a monomeric fluorophore with an absorption maximum at 587 nm and an emission maximum at 610 nm. mCherry has good photostability and is widely used for molecular markers and cell component positioning in biology.
Key features:
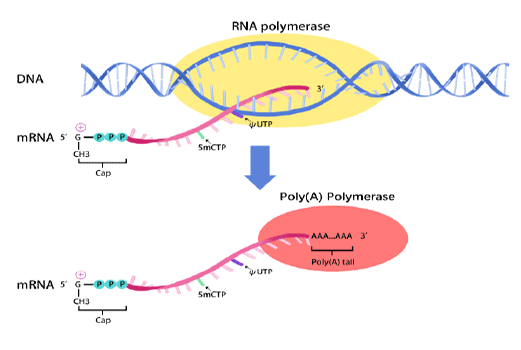
Cap1 Structure: The mRNA incorporates a Cap1 analog at the 5' end, ensuring high translation initiation efficiency, improved stability, and reduced innate immune recognition, leading to stronger and more sustained luciferase expression.
Modified Nucleotides: It contains 5mCTP and pseudo-UTP modified nucleotides, which decreases immunogenicity, enhances mRNA stability, and increases translational efficiency, resulting in more reliable and reproducible protein yield.
poly(A) tail: It contains an optimized poly(A) tail of approximately 100 nucleotides. This specific length is engineered to maximize transcript stability by resisting degradation and to synergize with the 5' cap for superior, sustained translation efficiency, ensuring robust protein yield in your applications.
| mRNA Length | 996 nucleotides | ||
| Concentration | 1.0 mg/mL | ||
| Buffer | 1 mM Sodium Citrate pH 6.4 | Storage | At or below -40°C |
| Application | Reporter Genes | Cap | Cap 1 |
- 1. Zhicheng Le, Jiang Qian, et al. "A versatile gemini amphiphile-based platform with STING-activating properties for efficient gene delivery into dendritic cells." Chemical Engineering Journal Volume 497, 1 October 2024, 154513
- 2. Roach, Arantxa, et al. "Kidney-Targeted mRNA Nanoparticles: Exploration of the mRNA Loading Capacity of a Polymeric Mesoscale Platform Employing Various Classes of Excipients." (2024). Biochemistry and Molecular Biology. 2.
- 3. Ina Guri-Lamce, Yara Alrokh, et al. "Lipid nanoparticles efficiently deliver the base editor ABE8e for COL7A1 correction in dystrophic epidermolysis bullosa fibroblasts in vitro." J Invest Dermatol. 2024 Apr 5:S0022-202X(24)00269-0. PMID: 38583743
- 4. Rachel Skelton , Arantxa Roach, et al. "Formulation of Lipid‑Free Polymeric Mesoscale Nanoparticles Encapsulating mRNA." Materials Horizons. PMID: 36163410
Quality Control & Datasheet
- View current batch:
Related Biological Data
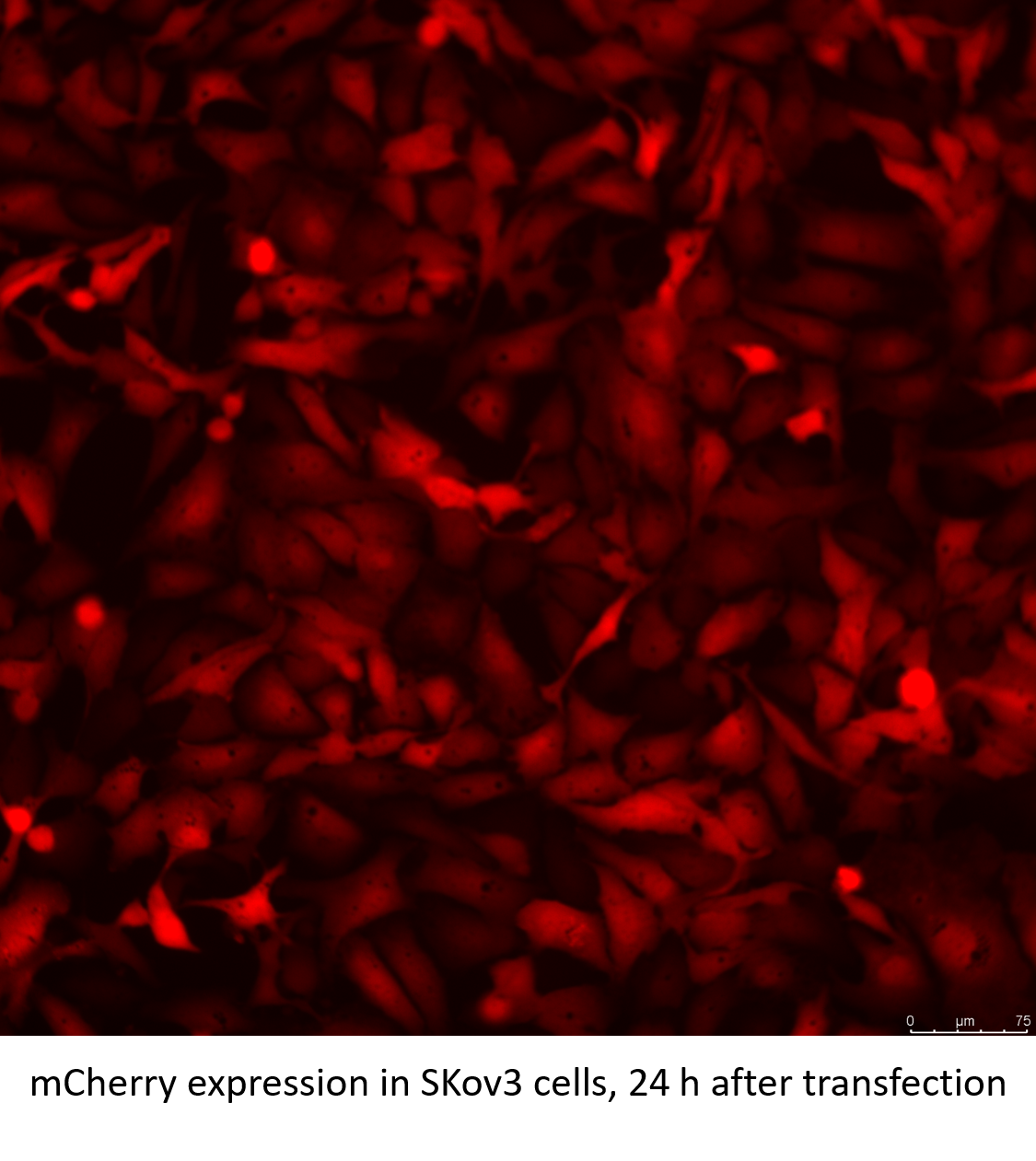
Related Biological Data
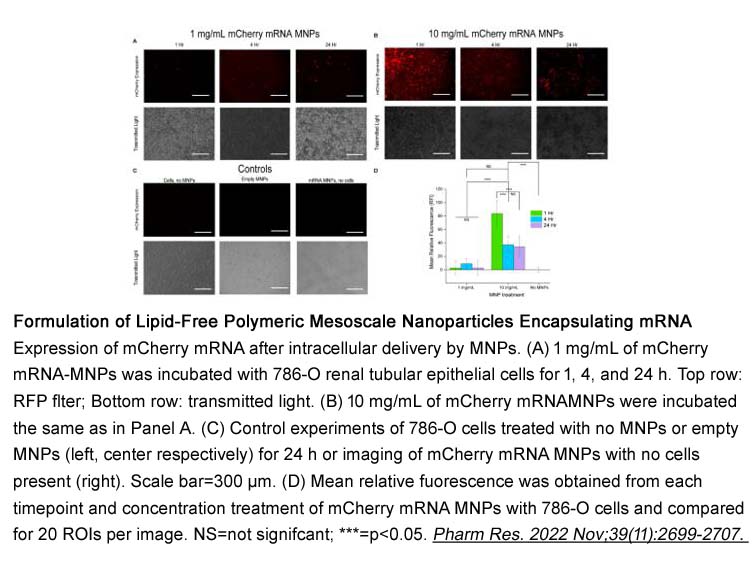
Related Biological Data
It can be based on your experimental goals:
- For Tracking Transfection and Translation Efficiency: APExBIO Reporter Gene mRNAs (e.g. EGFP, Firefly Luciferase mRNA) are commonly used to track transfection efficiency and protein expression duration; evaluate gene expression and cell viability; study mRNA localization and bio-distribution via in vivo imaging; optimize transfection conditions and validate LNP delivery system.
- For Gene Editing, Functional Studies and Gene Therapy Research: APExBIO offers various functional protein mRNAs, involving tumor suppressors (e.g. p53, PTEN), cytokines (e.g. IL-12, IL-10), gene-editing tools (e.g. spCas9, Cre Recombinase), gene replacement protein (e.g. EPO), and antigens (e.g. OVA, SARS-CoV-2 Spike).
- For Sustained Protein Expression: APExBIO Self-amplifying RNA (saRNA) and Circular RNA (circRNA) are recommended for applications requiring prolonged protein expression. saRNA enables lasting and strong protein expression at lower doses, while circRNA has enhanced structural stability and extended expression duration.
- Advanced Capping Technology: Utilizes Cap 1 structure (EZ Cap™ Cap) to achieve enhanced translation efficiency and minimizing activation of the host innate immune response. The capping efficiency can reach 90–99%.
- Diverse Modification Options: Provides a range of modified nucleotides, such as m1Ψ, 5-moUTP and Cy5-UTP, which reduce immunogenicity, improve mRNA stability, and maximize protein expression levels.
- Stringent Quality Control: Each batch undergoes rigorous quality assessment including capping efficiency, purity, integrity, and sterility to ensure batch-to-batch consistency.