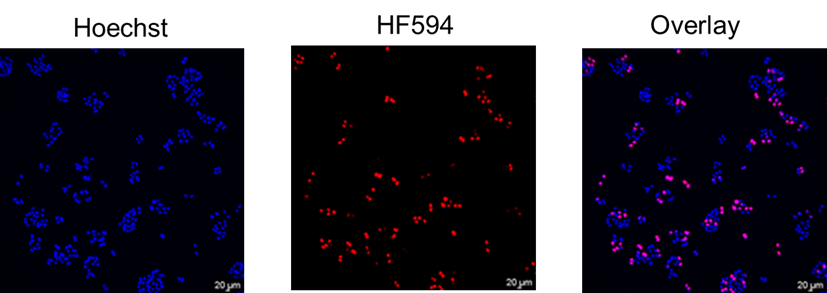
EdU Imaging Kits (HF594)
- 1. Yujie Ma, Runqiang Liu, et al. "Oleic acid inhibits the proliferation, invasion and migration of tongue squamous cell carcinoma cells under high glucose conditions through S100A9-HK2/PKM2-SOD2 axis." Tissue Cell. 2025 Jun 24:96:103026. PMID: 40582120
- 2. Yingjie Cai, Tong Guo, et al. "Alpha-Linolenic Acid from Zanthoxylum Seed Powder Regulates Fatty Acid Metabolism and Influences Meat Quality of Pekin Duck via the ADIPOQ/AMPK/CPT-1 Pathway." J Agric Food Chem. 2025 Jun 11;73(23):14651-14665. PMID: 40459020
- 3. Yan Hu, Chuntao Liu. "SIRT3-SUMO regulated Treg cell differentiation and asthma development by mediating N-glycosylation through the FAO pathway." Cell Biol Toxicol. 2025 Dec 13;41(1):164. PMID: 41388161
- 4. Tingting Qin, Zhangxu He, et al. "Taohe Chengqi decoction improves diabetic cognitive dysfunction by alleviating neural stem cell senescence through HIF1α-driven metabolic signaling." Phytomedicine. 2024 Dec:135:156219. PMID: 39520950
- 5. Ziqiao Yuan, Jiahui Meng, et al. "Formononetin Mitigates Liver Fibrosis via Promoting Hepatic Stellate Cell Senescence and Inhibiting EZH2/YAP Axis." J Agric Food Chem. 2024 Oct 3
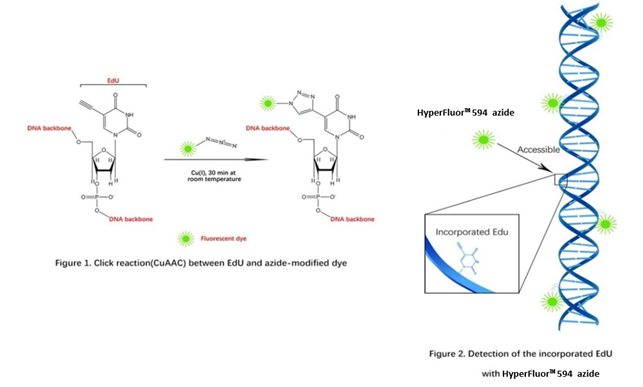
Measuring cell proliferation and cell cycle are a fundamental method to assess cell health, determine genotoxicity, and evaluate drug’s pharmacodynamic effect. The common method is measuring DNA synthesis directly. In previous experiments, there are several approaches such as the incorporation of radioactive nucleosides (3H-thymidine) or BrdU. Here, we introduce one new method, click chemistry - CuAAC (Copper-Catalyzed Azide-Alkyne Cycloaddition), and the use of this reaction in direct measurement of S-phase DNA synthesis in cell cycle. "Click Chemistry" is a term that was introduced by K. B. Sharpless in 2001, it’s a set of biocompatible small molecule reactions commonly used in bioconjugation, allowing the joining of substrates with specific biomolecules (such as a reporter molecule).
A nucleoside analog of thymidine, EdU (5-ethynyl-2’-deoxyuridine), can be incorporated into DNA strand during DNA synthesis. The alkynyl group of EdU is a biologically inert group that will undergo an extremely selective reaction with dye’s azido via a CuAAC reaction to afford an 1,2,3-triazole product. EdU and HyperFluor™ 594 azide possess biologically unique moieties to fluorescently label DNA of proliferating cells, producing low backgrounds and high detection sensitivities. This CuAAC reaction affords superior regioselectivity and quantitative transformation under extremely mild conditions. The reaction is a highly efficient and can be used in a quantitative manner via Flow Cytometry or fluorescence microscope.
EdU is an ideal alternative of BrdU since it’s independent of harsh DNA denaturing conditions (typically using HCl, heat, or digestion with DNase). In BrdU-based assays, it requires extra DNA denaturation to expose the BrdU to an anti-BrdU antibody which is huge compared to fluorescent dye used in EdU-based assays. Unlike bulky antibodies, alkyne and azide groups are very small in size, therefore HyperFluor™ 594 azide gain access to the DNA under standard aldehyde-based fixation and detergent permeabilization. Besides, BrdU-based method is time consuming and difficult to perform consistently, antibodies of BrdU can exhibit nonspecific binding. The harsh treatments necessary for this method can adversely affect sample integrity, cell morphology and image quality. Because of the mild reaction conditions in EdU-based assay, you can accurately determine cell proliferation while preserving cell morphology, DNA integrity, antigen binding sites. Preservation of DNA integrity allows for DNA staining, including staining with dyes used for cell cycle analysis.
HyperFluor™ 594 azide (Ex/Em: 590/617 nm)
| Components | 50-500 Tests | |
| EdU (Component A) | 5 mg | |
| HyperFluor™ 594 azide (Component B) | 1 vial | |
| DMSO (Component C) | 4 mL | |
| 10X EdU Reaction Buffer (Component D) | 4 mL | |
| CuSO4 (100 mM Aqueous Solution) (Component E) | 1 vial | |
| EdU Buffer Additive (Component F) | 400 mg | |
| Hoechst 33342 (10 mg/mL in Water) (Component G) | 35 μL | |
Store at -20ºC, keep dry and protected from light, stable for 1 year. | ||