Mirin
Mirin is a potent MRN complex inhibitor. Mirin inhibits Mre11-associated exonuclease activity, rather than alters DNA-binding or MRN complex formation. Moreover, mirin prevents MRN-dependent activation of ATM in response to DNA double-strand breaks, with an IC50 value of 12 μM. The MRN complex acts as a DNA damage sensor, responsible for maintaining genome stability during DNA replication, promoting homology-dependent DNA repair and activating ATM. The MRN-ATM pathway plays an essential role in sensing and signaling from DNA double-strand breaks.
Reference:
1. Dupré A, Boyer-Chatenet L, Sattler RM, et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nature Chemical Biology, 2008, 4(2): 119-125.
| Physical Appearance | A solid |
| Storage | Store at -20°C |
| M.Wt | 220.25 |
| Cas No. | 1198097-97-0 |
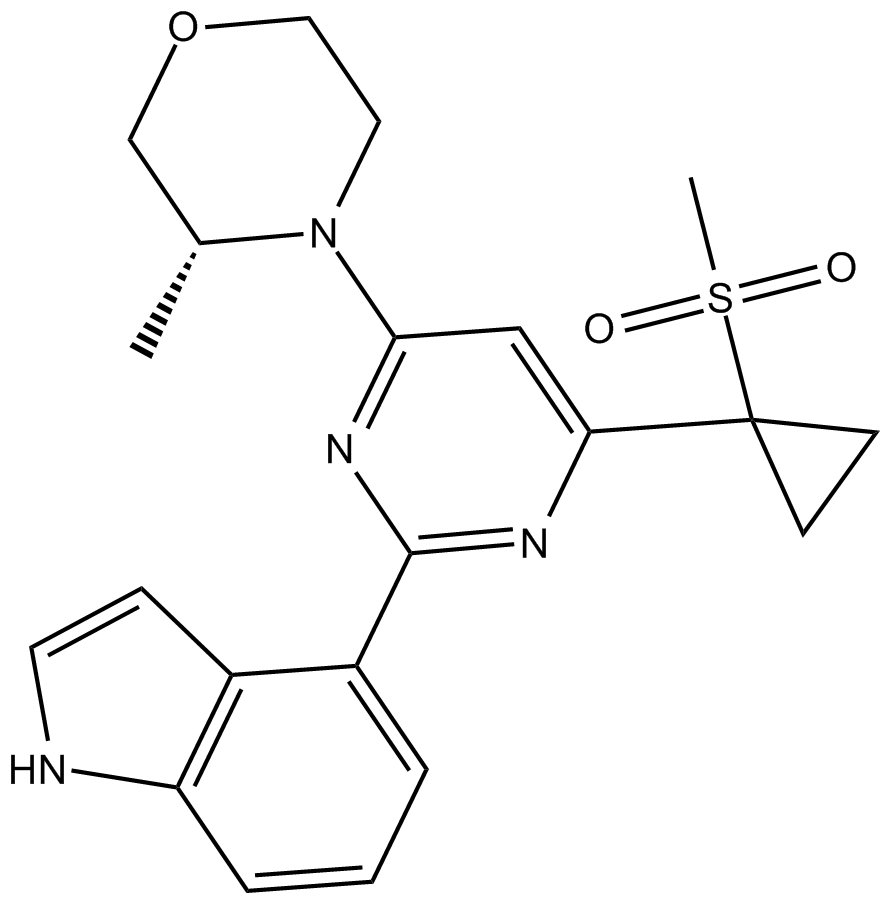
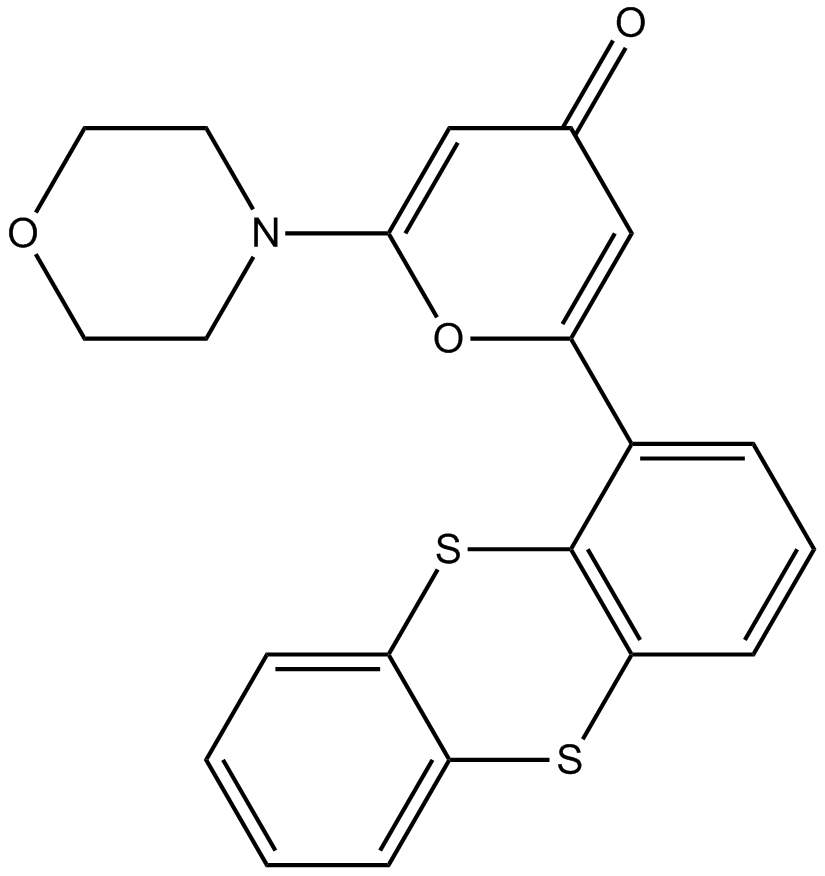
| Formula | C10H8N2O2S |
| Solubility | insoluble in EtOH; insoluble in H2O; ≥9.3 mg/mL in DMSO |
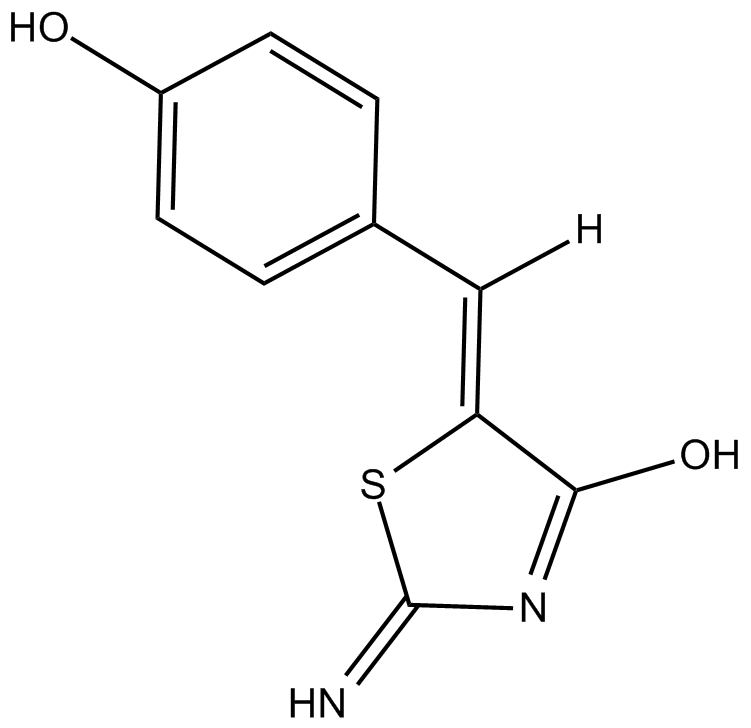
| Chemical Name | (Z)-5-(4-hydroxybenzylidene)-2-imino-2,5-dihydrothiazol-4-ol |
| SDF | Download SDF |
| Canonical SMILES | N=C(N=C1O)S/C\1=C\c(cc1)ccc1O |
| Shipping Condition | Small Molecules with Blue Ice, Modified Nucleotides with Dry Ice. |
| General tips | We do not recommend long-term storage for the solution, please use it up soon. |
| Cell experiment:[1] | |
|
Cell lines |
Human embryonic kidney (HEK)293 cells and TOSA4 cells |
|
Reaction Conditions |
10 ~ 100 μM mirin for 24 h incubation |
|
Applications |
In mammalian cells, mirin induced G2 arrest, abolished the radiation-induced G2/M checkpoint, and prevented homology-directed repair of DNA damage. |
|
Note |
The technical data provided above is for reference only. |
|
References: 1. Dupré A, Boyer-Chatenet L, Sattler RM, et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nature Chemical Biology, 2008, 4(2): 119-125. |
|
Quality Control & MSDS
- View current batch:
Chemical structure